Guided Tour of the Pathway Logic model of Mycobacterial Mycolic Acid Biosynthesis
Table of Contents
- Introduction to symbolic systems biology model
- Mycolic acid biosysnthesis
- BioCyc mycolic acid database view
- Pathway logic mycolic acid system model view
- Summary statistics of the pathway logic mycolic acid model
- Guided tour of the pathway logic mycolic acid model
- PLA overview of the model
- Getting information about graph nodes
- Analysis of TB-mycolic-acid graph
Introduction to symbolic systems biology model
Symbolic systems biology (SSB) is an approach to computational modeling of biological systems that builds on logical representations and knowledge bases. It uses the symbolic reasoning tools developed for analysis of complex hardware and software systems. To explain a biological process, one can have two views of the information collected in a knowledgebase: a database view and a system modeling view.
From the database view, a knowledgebase can be queried to find collections of stored objects that satisfy given conditions. For example, a query can request a list of all enzymes that catalyze a reaction in which ATP is a reactant.
From the system modeling view, objects from the knowledgebase can be assembled to represent a system state and relations (reactions) used to analyze the systems behavior. For example, how the state might change over time and what effects might result from the given starting state (signals, metabolites, secreted products produced).
Database queries can be used to generate system models and the results of model analysis can be used to add new information to the knowledgebase.
The database view is supported by BioCyc and the system model view is supported by Pathway Logic; both these systems are developed and coordinated at SRI International.
Mycolic acid biosysnthesis
The mycolic acid biosynthesis is a metabolic process that belongs to the bacteria, Mycobacterium tuberculosis (Mtb), a causative agent of the deadly disease, tuberculosis or TB.
To see an overview of mycolic acid biosynthesis and its relevance in drug discovery please click here.
Biocyc mycolic acid database view
The mycolic acid biosynthesis pathway was curated from the experimental literature and had been published on BioCyc. Please go here to view the pathway on BioCyc (For structural details: click on "More Detail" tab).
This pathway on BioCyc was acknowledged in the following:
- MetaCyc release note
- Nucleic Acids Research
The BIOPAX version is available here. We thank Malabika Sarker from SRI International for her contribution of the Mycobacterium tuberculosis mycolate biosynthesis pathway, by far the largest pathway in our database.
Pathway logic mycolic acid system model view
In Pathway Logic, we have curated the Mycolic acid biosynthesis pathway using the Maude system , a system based on rewriting logic. Here, the mycolic acid biosynthesis can be viewed for each single step starting from acetyl-CoA till the end product, mycolate. The model gives the reactions in terms of rules and the participants in forms of occurrences. Participants are either metabolites or enzymes that catalyze the reaction from one metabolite to the next with an increase in carbon numbers and structural complexities. Detail information on the enzymes and metabolites can be explored and the PubMed references for any reaction can be navigated. Moreover, the pathway can be queried and in silico experiments can be performed using the execution, search and model-checking tools of the Maude system. In silico experiments include - finding shorter biosynthesis or sub-pathways of choice; studying perturbations of the pathway either by gene knock outs (single and multiple) or mimicking metabolite-inhibition; comparing two related sub-pathways. In addition, pathways leading from different initial conditions can be generated automatically from collections of network elements to make dishes of your choice.
Thus the purpose of the mycolic acid Pathway Logic model is to:
- assist in hypothesis making
- help biologists to keep track of published experimental results
- arrange and rearrange the curated data into conclusions graphically
- provide accurate maps suitable for computational analysis
- computationally predict the effects of perturbations of network elements (genes encoding enzymes or metabolites).
The analysis capabilities of PLA complement those available using the BioCyc tool suite. For example, alternative paths in reaction networks are found, drug-enzyme interaction mechanisms have been modeled and the effects of inhibiting specific enzymes have been examined.
Summary statistics of the pathway logic mycolic acid model
| Number of Nodes | 546 |
| Number of Edges | 936 |
| Number of Rules | 272 |
| Number of Enzymes (A) | 36 |
| Number of Metabolites (B) | 238 |
| Number of Occurences (A+B) | 274 |
To see the table with the entire enzyme names and the corresponding rule numbers please click here.
Guided tour of the pathway logic mycolic acid model
The guided tour of the Pathway Logic model of Mycolic acid biosynthesis demonstrates the use of PLA for visual navigation as well as computational analysis of this pathway. To be self-contained, this guide provides a little introduction to the use of PLA. More details can be found in the SmallKB guide and the PLA reference manual .
In this guided tour, double click on a figure to see an enlarged version. Users need to single click to restore the figure to its original size.
PLA overview of the model
The graphical interface of the Pathway Logic tools, PLA, provides a graphical representation of the Pathway Logic model of TB-mycolic acid biosynthesis network and enables us to browse and analyze this network. Using PLA one can:
- display the network of all metabolic reactions of the pathway;
- formulate and submit queries to find subnets in pathways, for example, activating one enzyme without activating a second enzyme;
- compare subnets and/or pathways - what is shared, what is different;
- explore a network of rules to discover what is up/down stream of some component or rule and to see if they can be connected.
- Find knockouts - whose omission (enzymes or metabolites) prevents reaching a specified state.
Startup
If you haven't already started the Mycolic acid biosynthesis online demo, you can do so by clicking here and following the instructions.
When PLA starts up, a knowledge base manager window (title: PLA KBManager) appears in the upper left corner of your screen. The KBManager displays the TB (stands for tuberculosis) knowledge base (rule sets) that is highlighted in blue.
Screen shot of the PLA KBManager
Start by selecting a predefined dish: press the "Select Dish" button on the right side of the KBManager window. This produces a menu with two options: "Edit" and "PreDefined". Select (click on) "PreDefined". This produces a (sub)menu with a pre-defined dish "TB-mycolic-acid". Select "TB-mycolic-acid" (by clicking on the corresponding menu item). Shortly a PLA viewer window (titled InitialNet for TB-mycolic-acid (graph2)) will appear on your screen displaying the mycolic acid model (the network of reactions reachable starting from the occurrences in TB-mycolic-acid dish).
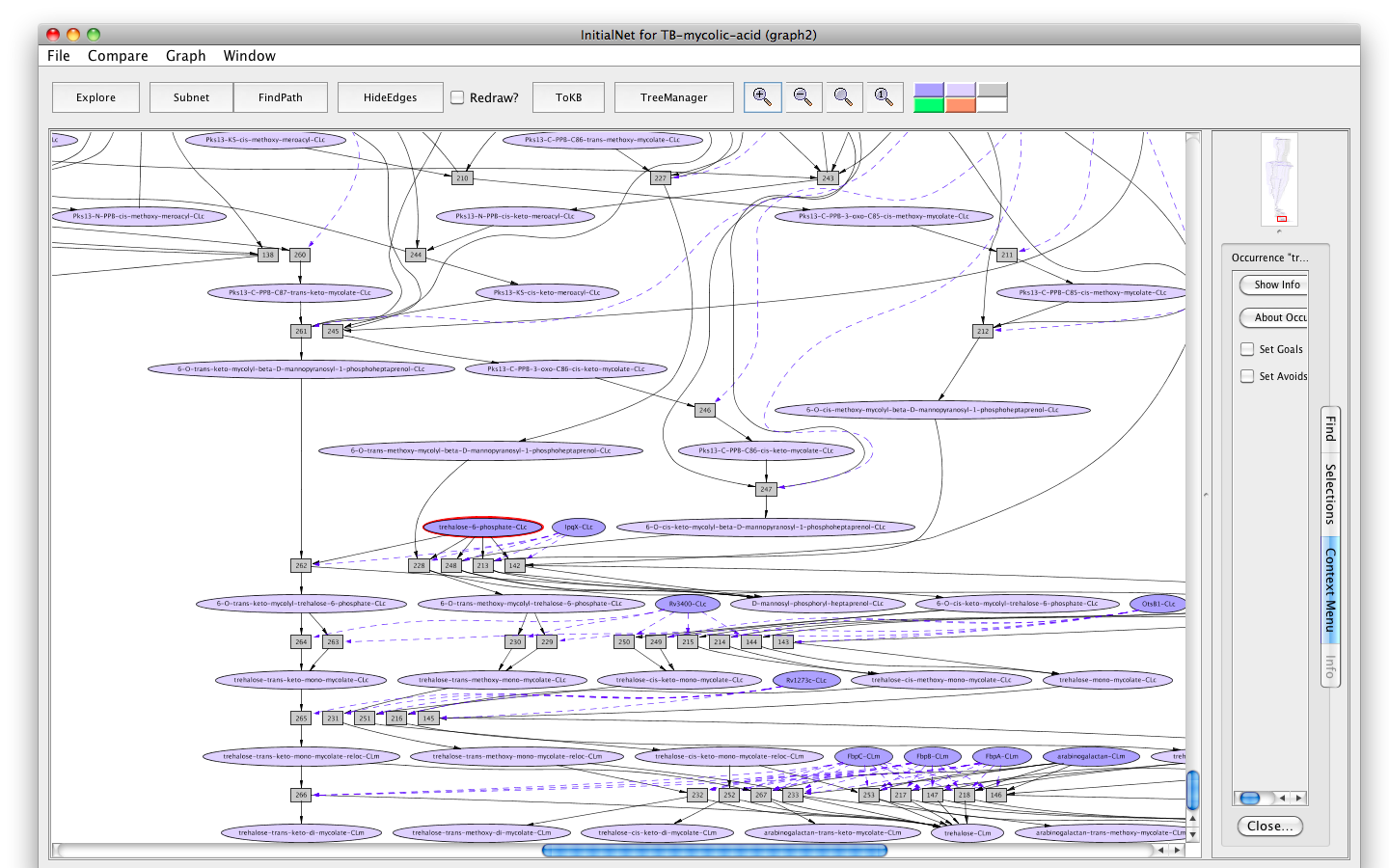
Screen shot of the TB-mycolic-acid graph
A network of reactions is displayed as a graph with two kinds of node. Ovals represent occurrences ---proteins (enzymes) or chemicals (metabolites) in a specific state and location. For example the oval labelled trehalose-6-phosphate-CLc represents the metabolite trehalose-6-phosphate located in the cytosol (-CLc).
Darker colored ovals represent occurrences in the initial state. Lighter colored ovals represent possible reachable states. Rectangles represent rules or transitions. The label in a rectangle is its (abbreviated) identifier in the knowledge base. Solid lines represent consumed or produced occurrences, depending on the direction of the arrow; dotted lines represent the enzymatic relation.
Abbreviations
We use the following abbreviations.
| Locations | ||
| CLm | ... | cell wall/cell membrane |
| CLc | ... | cytosol |
Lipid modifications
The modifications used here did not follow the standard Pathway Logic modifications since this pathway is a biosynthetic metabolic pathway and different from the rest of the pathways. The other pathways in Pathway Logic have types of modifications like "phos" for phosphorylation, "inhib" for inhibited etc. These modification terms were used for proteins. In mycolic acid pathway, we have proteins, lipoproteins and metabolites. The standard modifications for proteins are absent here since this is not a signaling pathway. To represent the lipoproteins we have put proteins as "occurrences" and added the lipid moieties as modifications. This is advantageous since proteins have their standard annotations available. As the list of modifications is too long, it has been put in a separate document. Please see here.
Navigating the TB-mycolic-acid graph
In the upper right of the PLA graph window is a thumbnail sketch of the full graph with a red rectangle delineating the part of the graph visible in the graph panel (on the left). Drag the rectangle around or click on different points in the thumbnail to change what is visible in the graph panel. In the lower right of the PLA graph window is an information pane with several tabs. The "Find" tab (the default) contains two lists: on the left a list of all occurrence nodes in the graph, and on the right a list of all rule nodes. You can use these lists to find a node of interest and center the view on that node. For example, type "in" in the text box above the occurrence list (under "Find Occurrence"). Tab completes "in" to "InhA-CLc", the first occurrence in the list whose label contains "in" as a prefix (ignoring case). Tab again to find the next match, "InhA:activated-Ethionamide-NADH-CLc". (Continued tabbing cycles through the matches.) Note that the matched label is highlighted in the occurrence list. Double click on the highlighted occurrence (or click the "Find" button at the bottom) to center the visible part of the graph on the occurrence of interest. You can also use the scroll bar on the right of the occurrence list to find the occurrence of interest and select it by clicking directly on the item in the list. Finding rules works in a similar manner. Some of the arrows lead to nodes not visible in the current view. To find out what is at the other end, click on the arrow. The information window (context tab) tells you what is the node at the other end. To go to the other end, either click on one of the buttons in the context window or double-click on the arrow.
Getting information about graph nodes
Clicking on a node displays information options in the Context tab of the information pane.- Click on the oval labelled InhA-CLc. The information panel (on the right) presents options for finding out about this occurrence. The button labelled Show info displays information for debugging.
- Click on the button labelled About Occurrence. Now the panel displays information about InhA: its KEGG id, SwissiProt id, EC number, Entrez-Gene id, Protein bank id, TubercuList id and synonym(s). All the ids are linked to the corresponding web pages.
- Click on the SwissProt link. A browser window with the requested page will appear. (You can close the browser window when you are done looking at the page, or keep it around. Its independent of PLA.)
- Similar information is available for metabolites. For example, click on the oval labelled malonyl-CoA-CLc. The KEGG compound id, the PubChem id and the synonyms are displayed. The ids are hyperlinked.
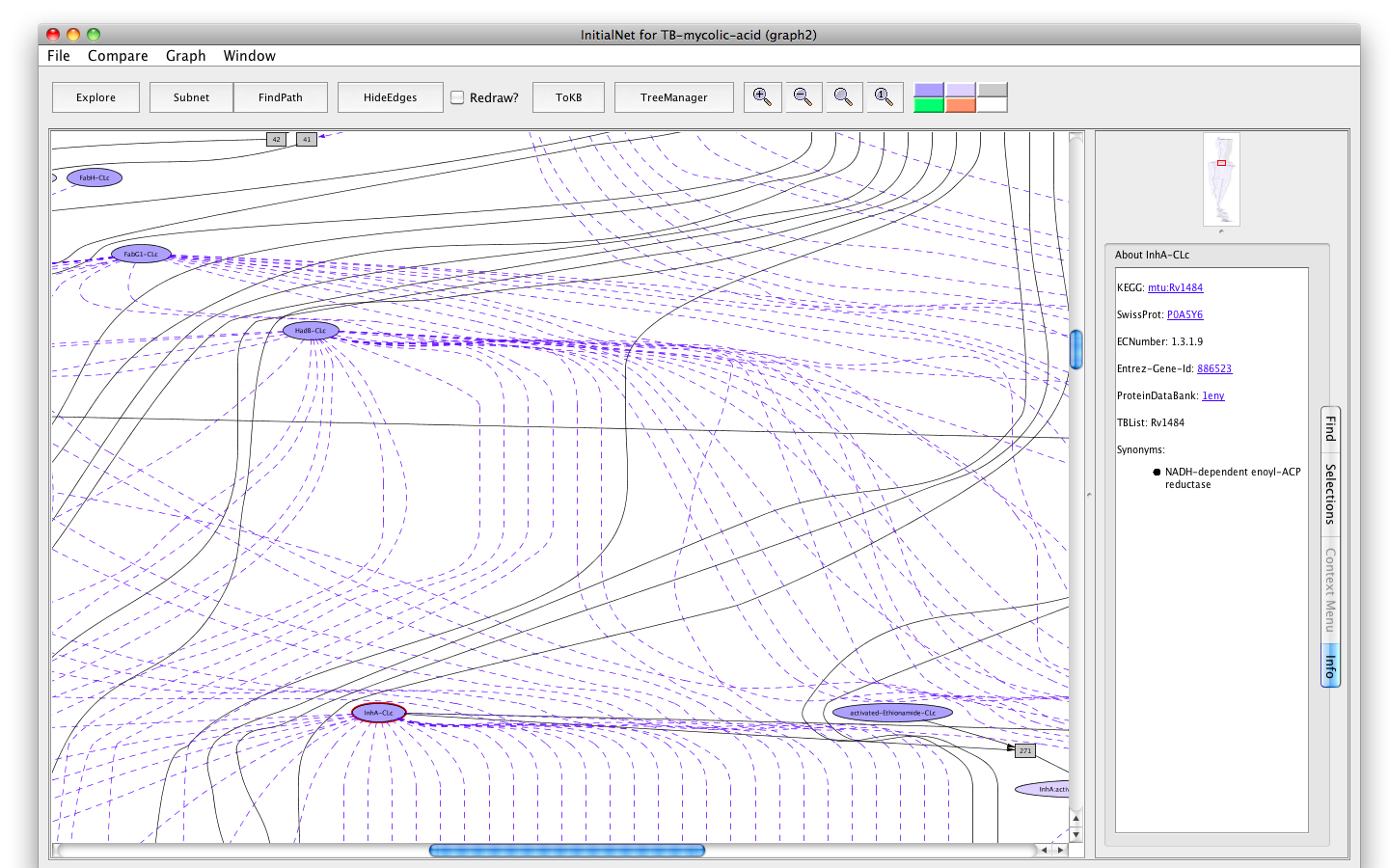
Screenshot of the enzyme InhA-occurrence tab info
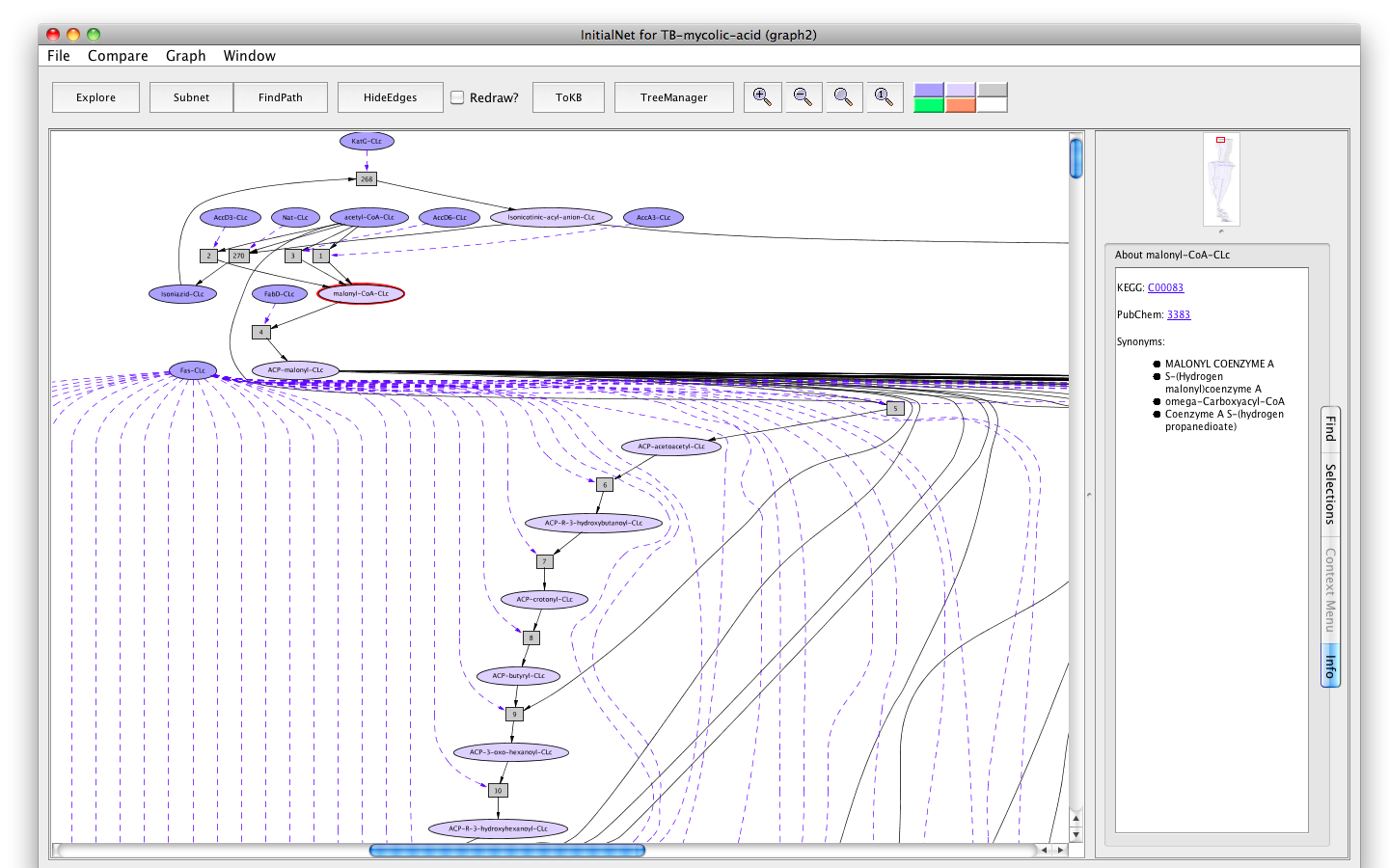
Screenshot of the metabolite malonyl-CoA-occurrence tab info
- Click on rule 267 (the rectangle labelled "267"). The information panel presents options for finding out about rule 267. The button labelled Show info displays information for debugging; the button labelled Show Maude Rule displays the rewrite rule corresponding to the biochemical reaction in Maude syntax.
- Click on the button labelled Show Evidence. You will find a Pubmed link to the paper used in curating rule 267.
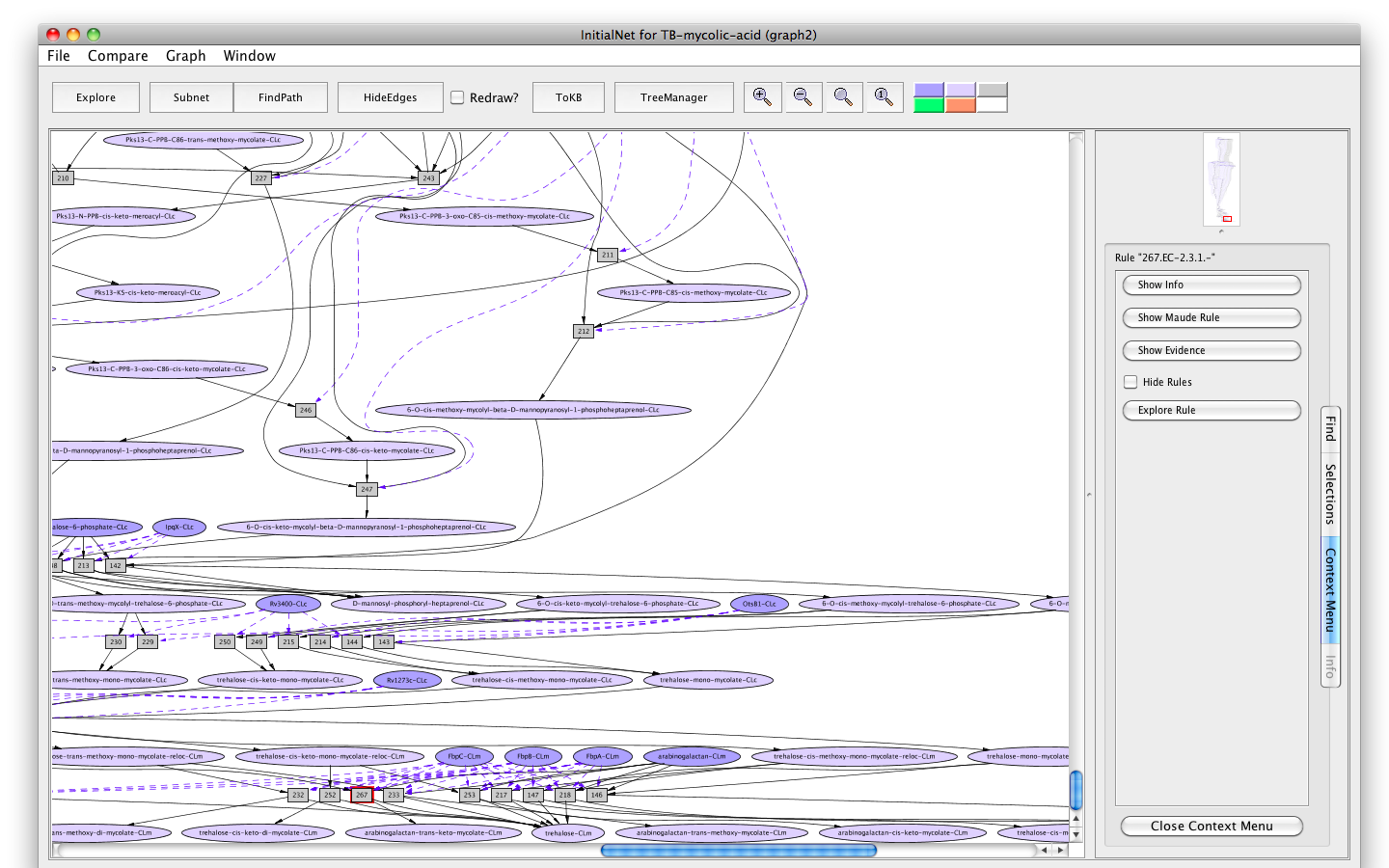
Screenshot of the information panel for rule "267"
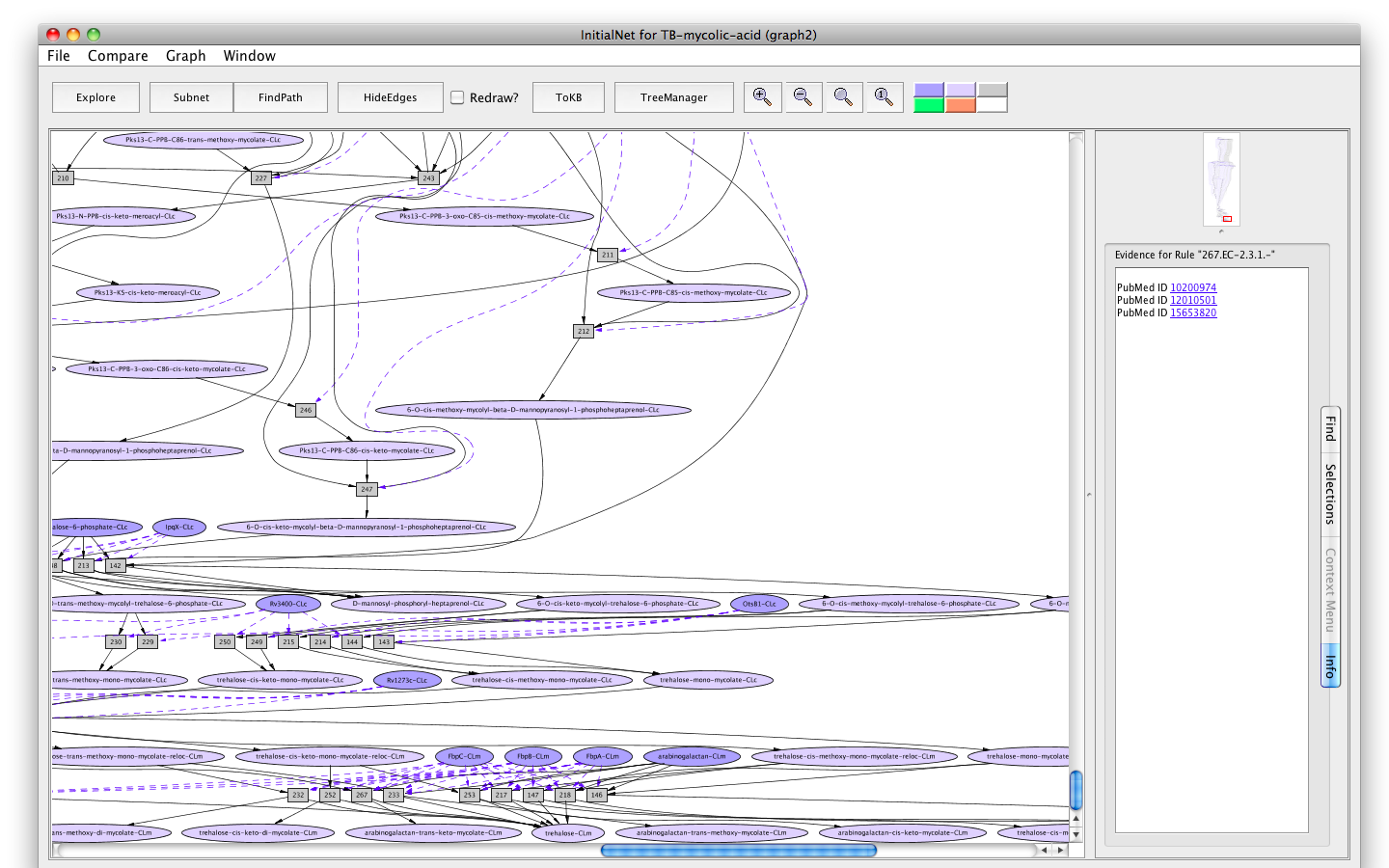
Screenshot of the Evidence of rule "267"
Analysis of TB-mycolic-acid graph
In the following sections, we show how meaningful results were derived from the analysis of TB-mycolic-acid network. This includes finding subnets, comparing them, in silico knockout studies and drug-enzyme interaction. Specifically, we have examined the following:
- Synthesis of an acyl carrier protein (ACP)-linked fatty acid intermediate, ACP-malonyl as an example.
- In silico single knock-out of a critical enzyme - MmaA4, a cyclopropanation enzyme.
- In silico inhibition of an apparently important metabolite - 2-carboxyl-cerotoyl-CoA.
- Activation of an anti-TB drug, Isoniazid, by KatG; inhibition of enzyme target InhA by both Isoniazid and ethionamide; the deactivation of Isoniazid by Nat.
- Comparison of synthesis for trans and cis forms of trehalose-keto-di-mycolate.
For simplicity, cofactors like ATP, CoA, H+, Mn2+, NADH, NADPH and the protein ACP have been hidden. The list can be viewed by "Show unused" button from the "Graph" menu.
Find "Subnet" and "FindPath" using "Goals"
A "Subnet" or a pathway is a collection of rules assembled in response to a query. A query specifies goals --- occurrences that should be present, i.e. states and locations of enzymes or metabolites. It may also specify occurrences or rules to be avoided. The execution of a subnet leads from the model's initial state to a state satisfying the goals, while not passing through any states containing occurrences to be avoided, or using any rules to be avoided. For the tour we will find one subnet, the one that synthesizes the ACP-linked fatty acid intermediate ACP-malonyl.
Generation of ACP-malonyl pathway:
- Make ACP-malonyl-CLc visible in the graph by using the "Find" tab.
- Click on the occurrence ACP-malonyl-CLc.
- Click on the "Set Goals" button in the information panel. The node will turn green.
- Press the button labeled "Subnet" in the tool bar. Shortly a new window will appear displaying a graph "Subnet of TB-mycolic-acid (graph 4)" representing a subnet leading to the synthesis of ACP-malonyl.
- You will see a new button in the tool bar labelled "In Context". Press this button and you will see the subnet in the context of the original graph (context nodes not in the subnet are white). The button is now labelled " DOT layout". Press again to get back to the original rendering.
Note that whenever you find and analyze pathways or subnets, make sure to reset the goal selection: either click the Set goals/ avoids button for highlighted proteins if it is still visible, or select the "Selections tab" (on the right of the information panel) and click the button labeled Reset All Selections at the bottom. This will prevent unwanted pathways or subnets from showing up in your desired pathway/ subnet analysis.
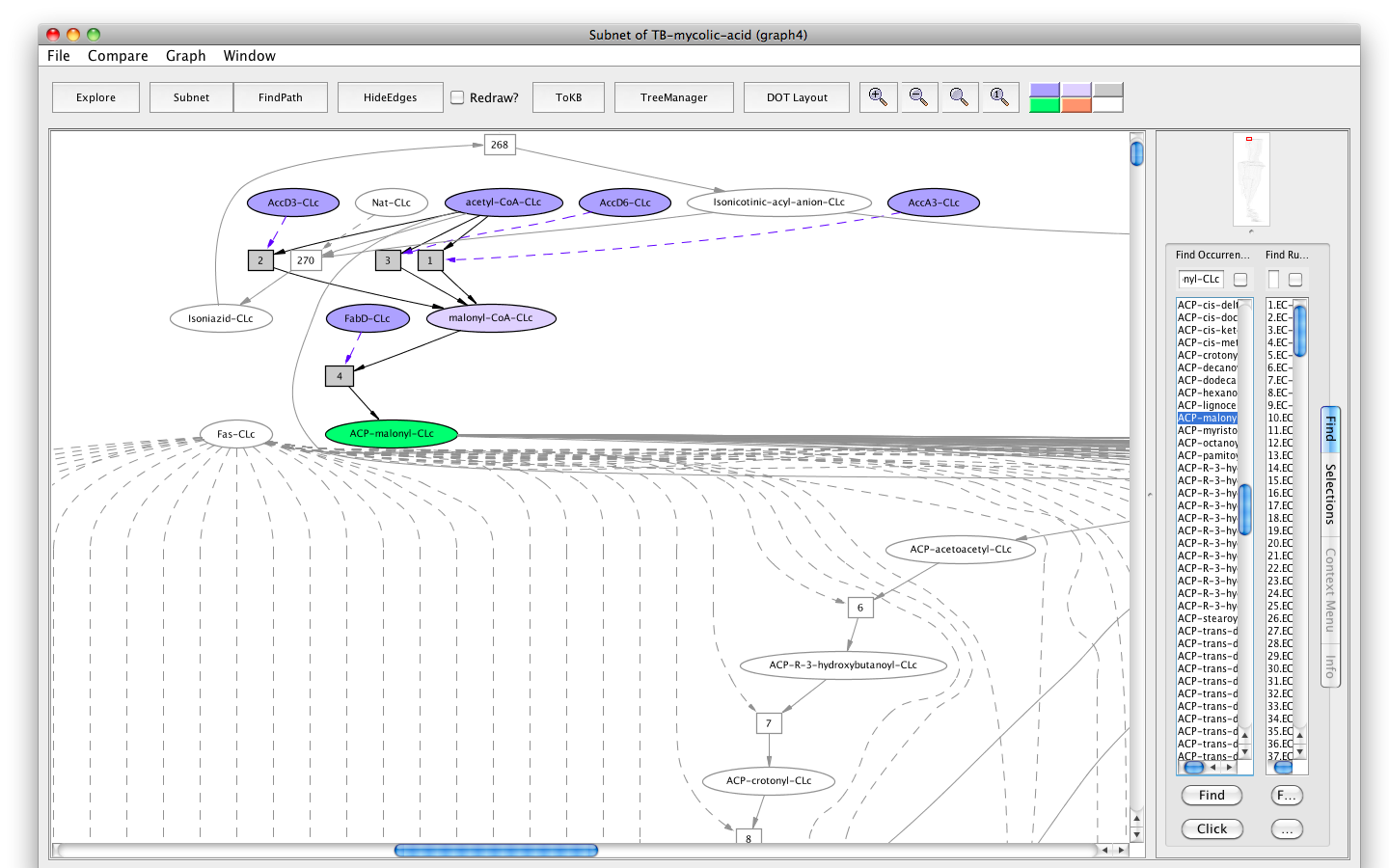
Screenshot of the "Subnet" with ACP-malonyl-CLc synthesis as goal in context of the original graph
"FindPath" will find one path, usually the shortest, among all possible paths for a specific goal. It is possible to get this path only when graph4 or the subnet is a network containing more than one route to get to the goal. Interestingly, the huge mycolic acid biosynthesis presented here contains all non-redundant pathways and therefore for any particular goal there are no alternative pathways. For this reason the "FindPath" option will not work here.
Find in silico knock-out for enzymes using "Avoids"
Let us see the effect of in silico single knock-out of an enzyme of Mtb. This enzyme is MmaA4, a methyltransferase. It is required for introducing the distal-branch methyl group in the formation of trans-oxygenated methoxy and keto meroacids [ 15653820 ].
- Return to the original graph window - InitialNet for TB-mycolic-acid.
- Reset the goal selection: select the "Selections tab" (on the right of the information panel) and click the button labeled Reset All Selections at the bottom.
- Click on the occurrence MmaA4-CLc.
- Click on the " Set Avoids" button in the information panel. The node will turn peach.
- Press the button labelled "Subnet" in the tool bar. Shortly a new window will appear displaying a graph representing a subnet without MmaA4-CLc and nodes influenced by it.
- Press the "In Context" button and you will see the subnet in the context of the original graph (context nodes not in the subnet are white).
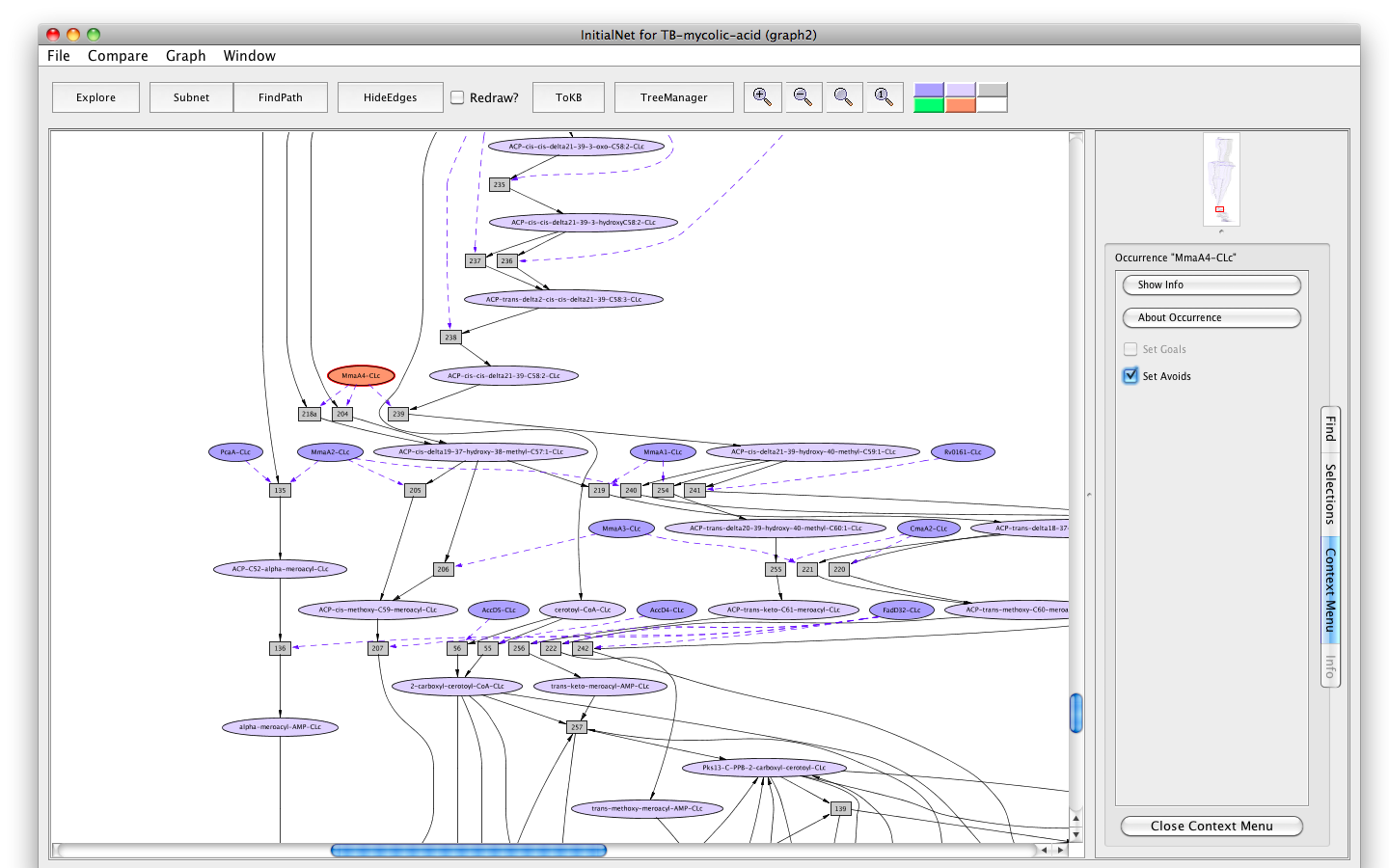
Screenshot of the setting "MmaA4" as "Avoids"
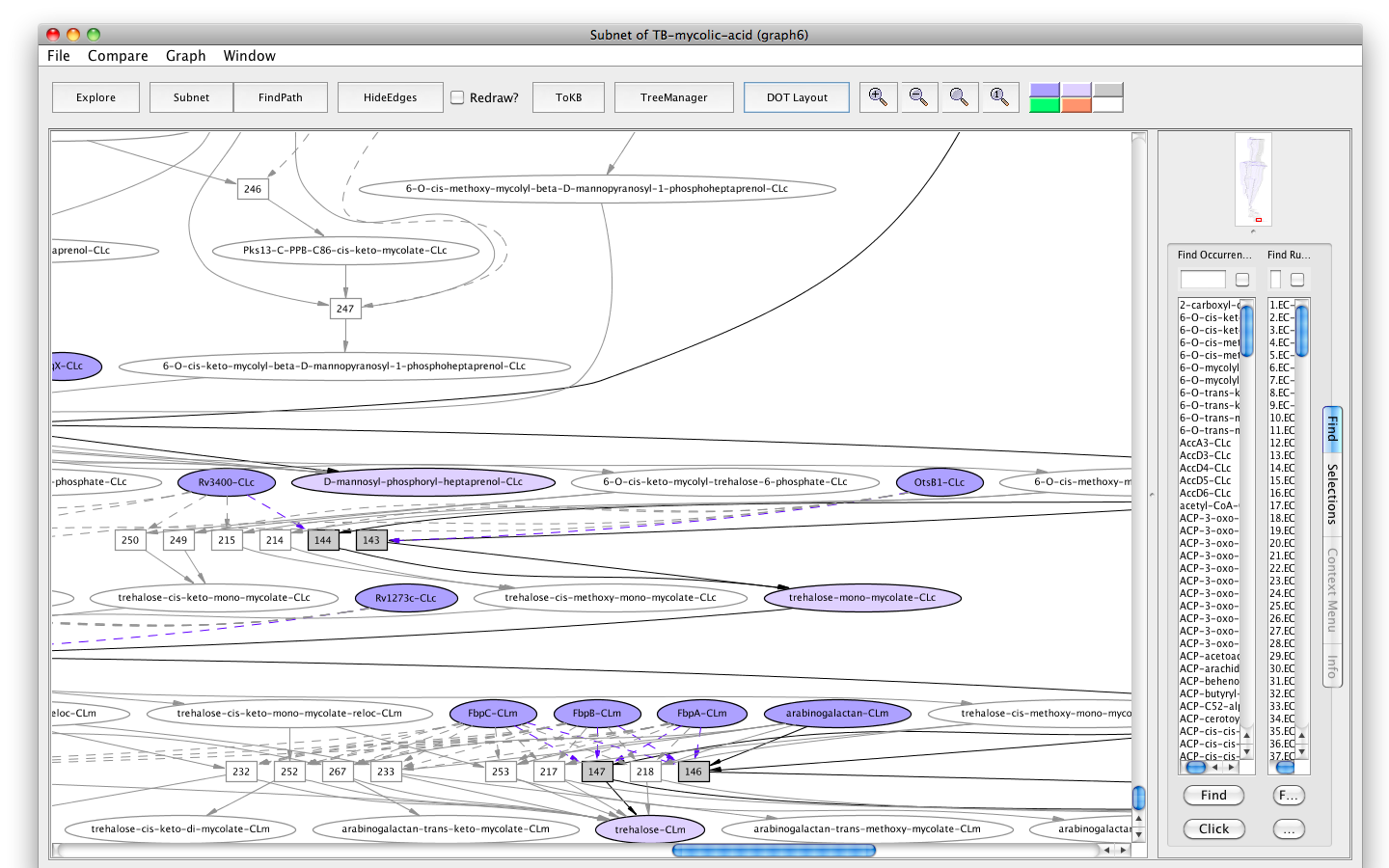
Screenshot of the "Subnet" without "MmaA4" in context of the original graph
Here we can see an interesting outcome. The synthesis of eight among ten final products, trehalose-cis/trans-mono/di-methoxy-mycolate, trehalose-cis/trans-mono/di-keto-mycolate, arabinan-cis/trans-methoxy-mycolate, and arabinan-cis/trans-keto-mycolate were inhibited. The synthesis of the other two final products, arabinan-(alpha)-mycolate and trehalose-(alpha)-mono/di-mycolate is rather unaffected.
Now, how does this happen?
- For the non-oxygenated alpha-mycolates, Mtb needs MmaA2 and PcaA, the cyclopropane synthases.
- For cis-methoxy-mycolate branch, Mtb needs MmaA4 and MmaA3 as methyltransferase and MmaA2 and CmaA2 as cyclopropane synthase.
- For trans-methoxy-mycolate branch, Mtb needs MmaA4, MmaA1 and MmaA3 as methyltransferase and CmaA2 as cyclopropane synthase.
- For cis-keto-mycolate branch, Mtb needs MmaA4 as methyltransferase, an unknown redox enzyme, and MmaA2 as cyclopropane synthase.
- For trans-keto-mycolate branch, Mtb needs MmaA4 and MmaA1 as methyltransferase, an unknown redox enzyme and CmaA2 as cyclopropane synthase.
of all the oxygenated mycolates (methoxy and keto) and not the non-oxygenated alpha-mycolates. The mmaA4 mutant further supports this analysis outcome. Mutant of mmaA4 synthesized only alpha-mycolic acid and not the oxygenated mycolate [ 12473649 , 10844652 ]. Genetic complementation was however found to restore the production of oxygenated mycolate [ 12473649 ]. Furthermore, a mutant strain of Mtb with an inactivated mmaA4 gene had profound alterations in its envelope permeability and is attenuated in mice [ 10844652 ]. Among all these enzymes, PcaA cyclopropane synthase and MmaA3 methyltransferase are found to be essential in vivo [ 14569030 ].
Deletion of the proximal cyclopropane ring of alpha-mycolic acid or of methoxy- and keto-mycolates in Mtb leads to a significant attenuation in growth of the two mutants in the mouse model of infection. A deletion of the keto-mycolates leads to restricted growth of this mutant in macrophages. Thus, the fine structure of mycolic acids is associated with virulence of Mtb [ 15653820 ].Find inhibition of metabolites using "Avoids"
Metabolite analogs or mimics are structurally similar to the microbial metabolites and interfere with the functions of the corresponding native metabolites. In this way, they act as selective inhibitors of microbial growth when the metabolites are essential for survival [ 13008406 ]. Examples of commercial antimicrobials that are anti-metabolites include Sulfamethoxazole, Trimethoprim, D-cycloserine and many more. The mimic strategy has successfully identified several compounds with in vitro activity against TB [ 21285434 ]. Recently, we identified ten metabolites as potential candidates for the development of novel antibiotics for TB through an in silico pathway analysis.
Now, let us see the effect of in silico inhibition of an Mtb metabolite 2-carboxyl-cerotoyl-CoA-CLc.
- Return to the original graph window - InitialNet for TB-mycolic-acid.
- Reset the goal selection: select the "Selections tab" (on the right of the information panel) and click the button labeled Reset All Selections at the bottom.
- Click on the occurrence 2-carboxyl-cerotoyl-CoA-CLc.
- Click on the " Set Avoids" button in the information panel. The node will turn peach.
- Press the button labelled "Subnet" in the tool bar. Shortly a new window will appear displaying a graph representing a subnet without 2-carboxyl-cerotoyl-CoA-CLc and nodes influenced by it.
- Press the "In Context" button and you will see the subnet in the context of the original graph (context nodes not in the subnet are white).
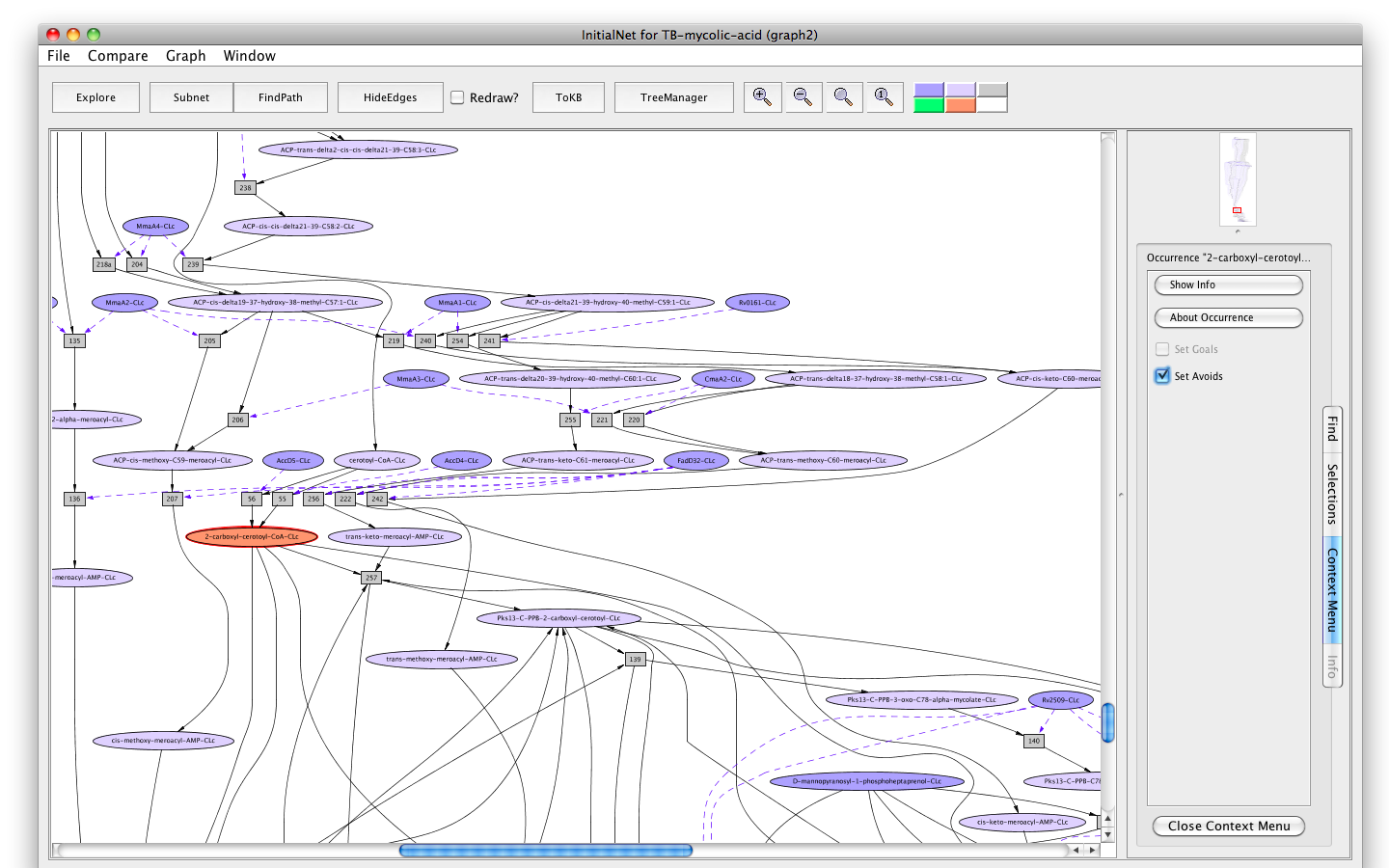
Screenshot of the setting "2-carboxyl-cerotoyl-CoA-CLc" as "Avoids"
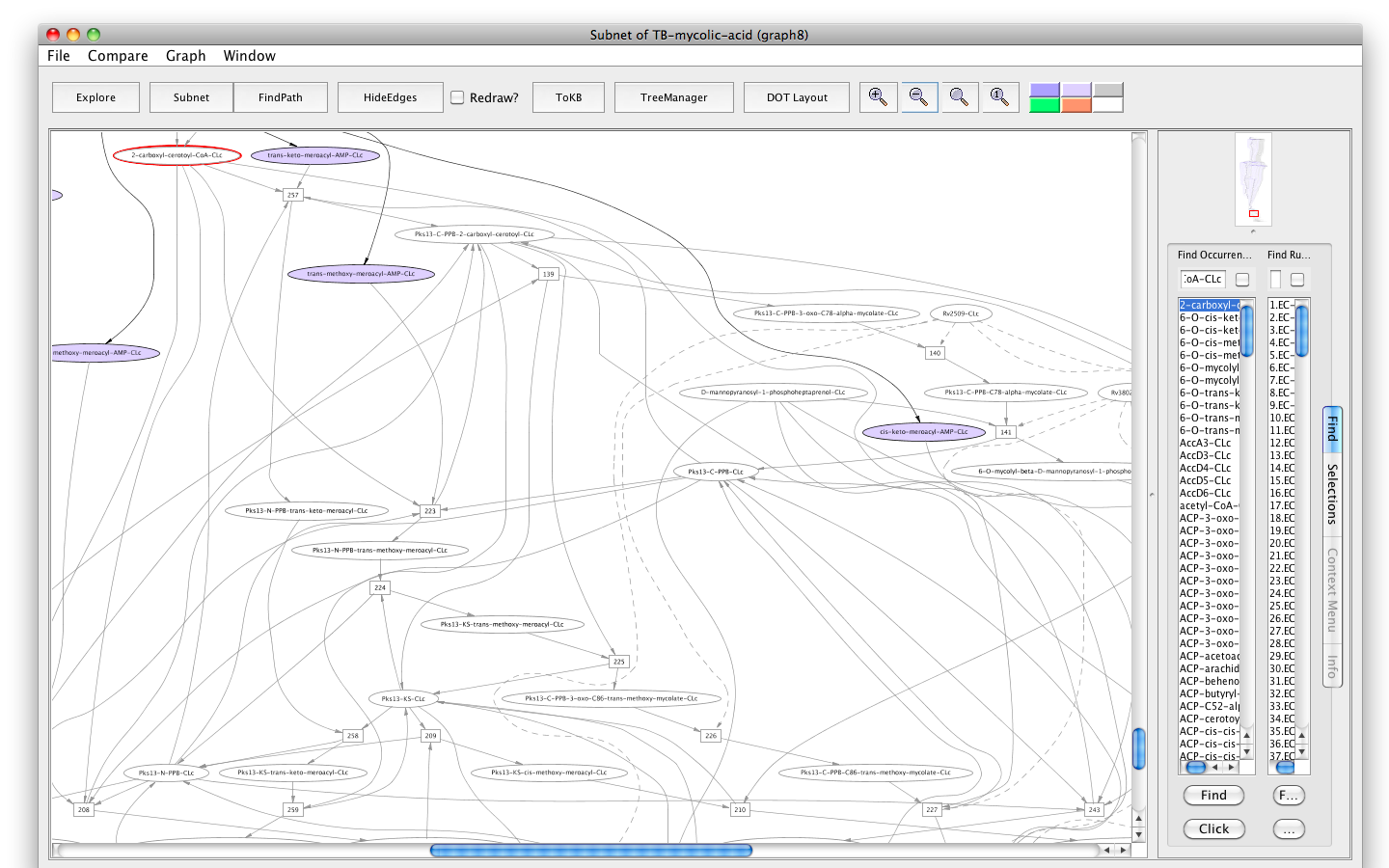
Screenshot of the "Subnet" without "2-carboxyl-cerotoyl-CoA-CLc" in context of the original graph
The result shows that a big chunk of the pathways that lead to the final products are non-functional. Therefore, the metabolite, 2-carboxyl-cerotoyl-CoA, seems to be critical. Experiments are needed to confirm if its inhibition can prevent Mtb growth in vitro.
Find "drug-enzyme" interactions
The Mycolic acid pathway curated on BioCyc platform was not able to show the anti-TB drug's action on target enzymes on this pathway. This is a special feature that we nicely represent in the Pathway Logic model. We show the activation and deactivation of the drug Isoniazid by other enzymes as well as the inhibition of enzyme InhA of the mycolic acid pathway by two drugs Isoniazid and Ethionamide.
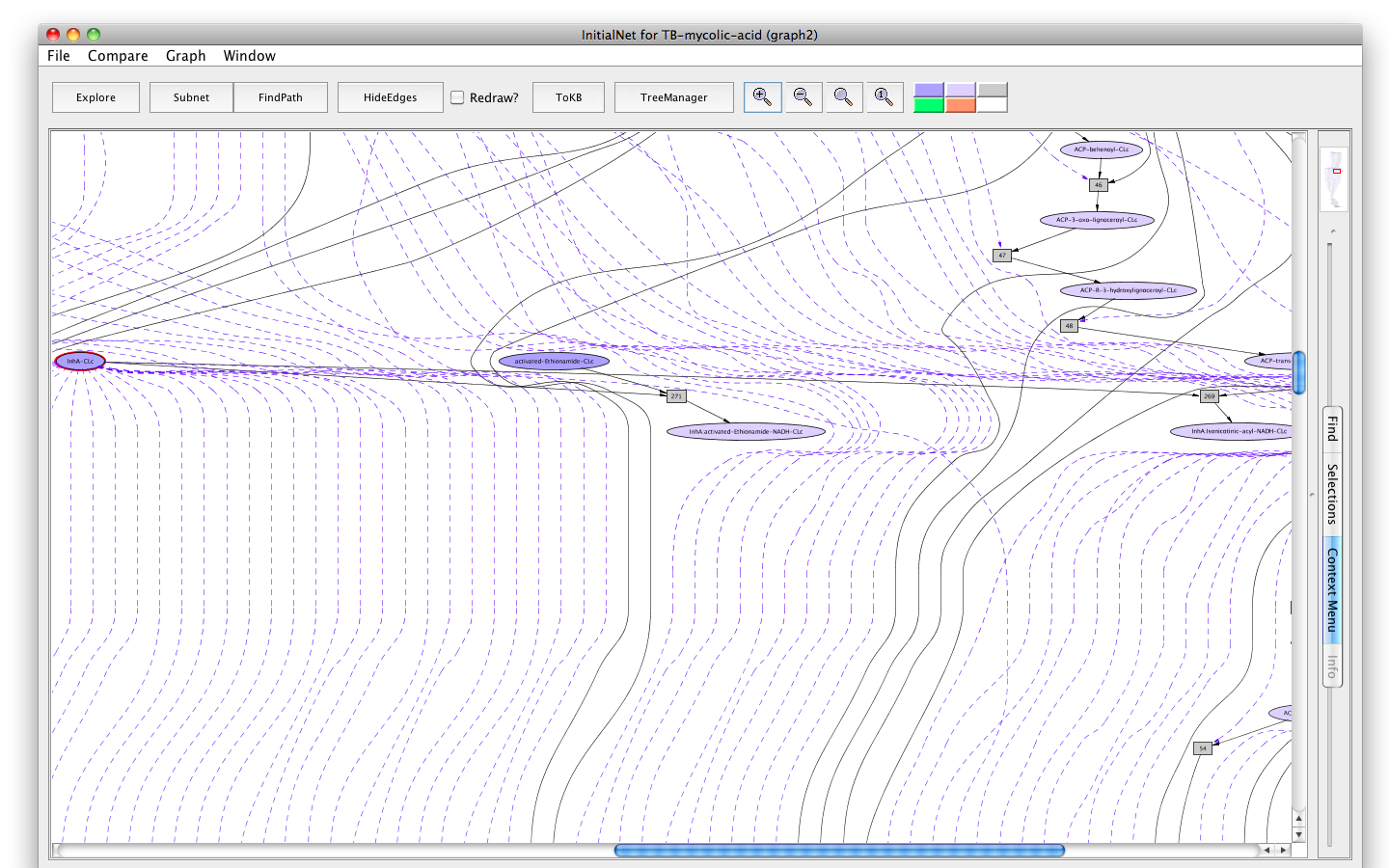
Screenshot of the interaction of drugs with InhA
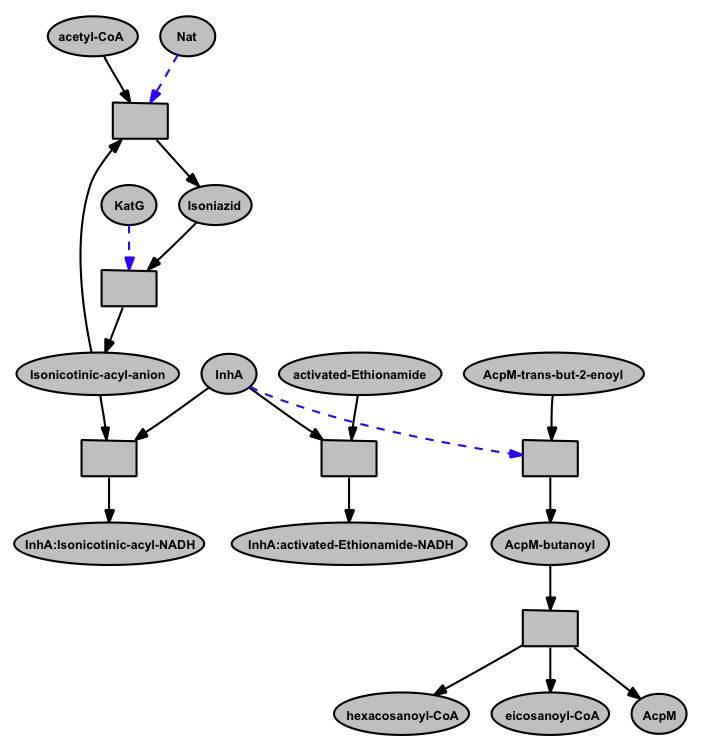
A closer view of the model
InhA (EC 1.3.1.9, NADH-dependent enoyl-ACP reductase), involved in mycolic acid synthesis, is a common target of Isoniazid and Ethionamide, a first line and second-line anti-TB drug respectively [ 12164478 ]. The network shows the mechanism of Isoniazid drug activation to Isonicotinic-acyl anion by KatG (dashed arrow) [ 20554537 ]; targeting InhA by activated Isoniazid through covalent attachment of the Isonicotinic-acyl anion to NADH [ 9417034 ]; inactivation of Isoniazid by Nat (Arylamine N-acetyltransferase (dashed arrow) [ 15722451 ]; targeting InhA by activated-Ethionamide [ 8284673 ], all in the context of the mycolic acid biosynthesis. These are represented in form of rules in the network. For simplicity the rule identifiers are kept hidden in the closer view.
When InhA is bound the activated drugs, the downstream reactions involving InhA get affected. That's how these drugs inhibit the synthesis of mycolic acid biosynthesis in Mtb by targeting InhA. However, the inability to activate the drugs (example, mutation in KatG) and/or the deactivation of Isoniazid by Nat can lead to drug resistance.
Compare two subnets using "Compare"
We first generate two pathways that are of importance to us and find components that participate in both these pathways. Then, we test the importance of these common components by inhibiting it in a larger network by identifying them as critical components or as choke points.
- Synthesis of trehalose-trans-keto-di-mycolate: For this, find a subnet with trehalose-trans-keto-di-mycolate as goal.
- Synthesis of trehalose-cis-keto-di-mycolate: For this, find a subnet with trehalose-cis-keto-di-mycolate as goal.
Now we can compare the synthesis of trehalose-trans-keto-di-mycolate and that of trehalose-cis-keto-di-mycolate. In the new pathway window (the last available subnet) click on the menu labeled "Compare". The window hierarchy will be displayed (gray names are not comparable graphs). Choose Subnet of TB-mycolic-acid, graph 4. A new graph viewer window will appear displaying the comparison graph (graph 8). Nodes that belong to both graphs are colored peach, those only in the starting graph (the pathway to trehalose-trans-keto-di-mycolate-CLm) are colored purple, and those only in the compare to graph (the pathway to trehalose-cis-keto-di-mycolate-CLm) are colored cyan. (You can get this information from the color key at the right of the tool bar by mousing over the color of interest.) You can see the comparison graph in the context of the common parent (InitialNet for TB-mycolic-acid) by clicking on the "In Context" button.
In the comparison graph (graph 8), we see the major portion of the pathway, up to the synthesis of ACP - cis-delta21-39-hydroxy-40-methyl-C59:1 (peach colored), is common to both the synthesis pathways. However, the involvement of the different cyclopropanation enzymes, namely MmaA2 and MmaA1 plus CmaA2 divides the final paths to trans-keto- and cis-keto-mycoates respectively.
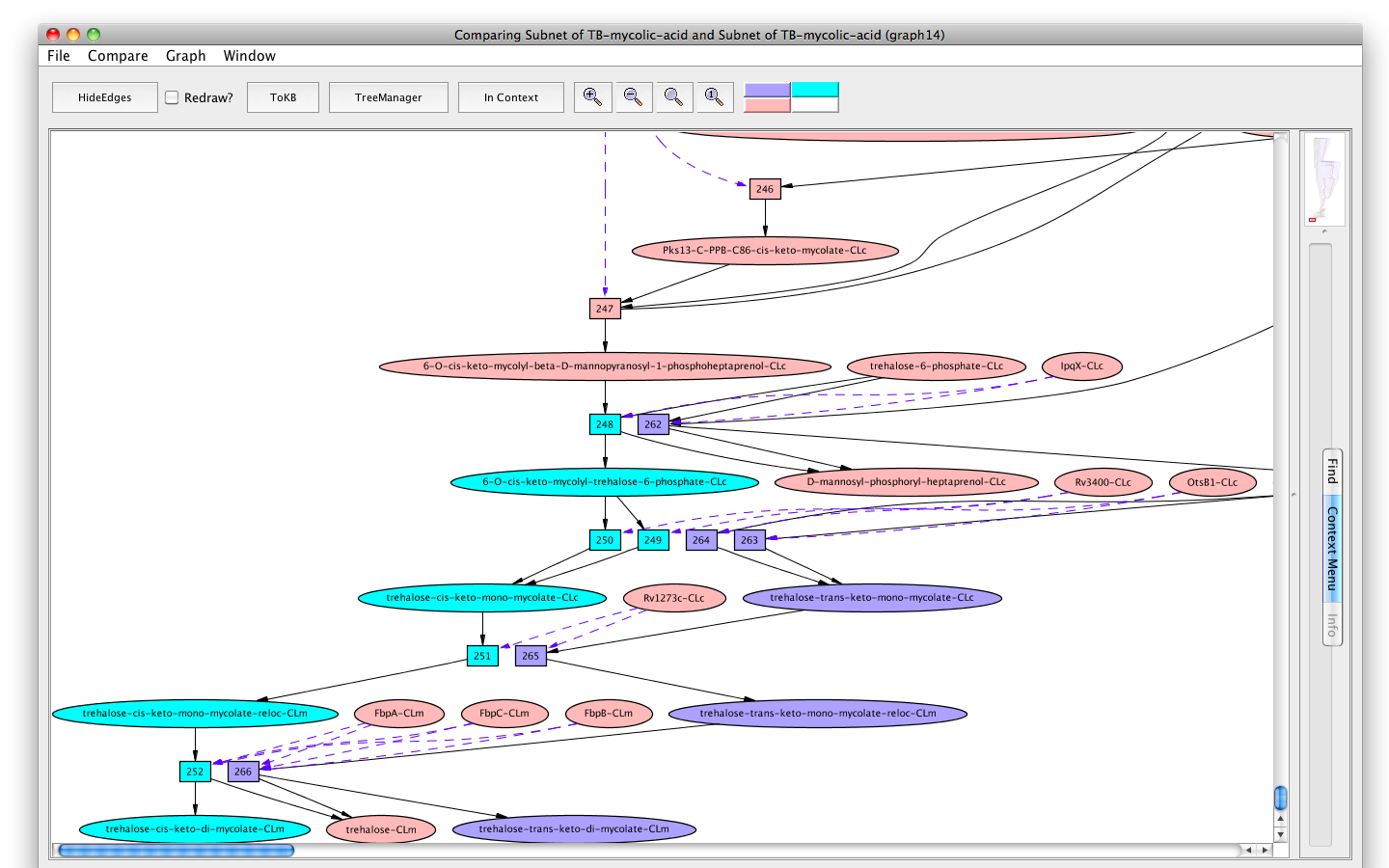
Screenshot of the comparison graph of synthesis of trehalose-trans/cis-keto-di-mycolate