STM7: A Pathway Logic Model of Intracellular Signal Transduction
Introduction
STM7 is a formal knowledge base containing information about the changes that occur in the proteins inside a cell in response to exposure to receptor ligands, chemicals, or various stresses. Ultimately, we intend to link these responses to changes in gene expression and cellular morphological phenotypes such as induction of proliferation and differentiation. So far we have been able to produce hypothetical pathways leading to the activation of a few transcription factors and epigenetic regulators. PLA provides a graphical representation of the STM7 reaction network and the ability to browse and analyze this network.
The STM7 model is a graph of the changes that occur when peptides, chemicals, or stresses are added to cultured cells. For simplification the graph is divided into 32 maps -- each representing one stimuli. These maps are called "dishes" because they describe an initial state which corresponds to the state of the cells in a culture dish at the start of an experiment, plus a stimuli. The base initial state in each of these dishes is the same--the cells are untreated and quiescent. Each dish is obtained by adding to the base state one ligand or stimulus and it's associated receptor(s) if applicable. The currently available stimuli are: Adriamycin, Anisomycin, Bleomycin, anti-Cd40, Csf1, Egf, Etoposide, Hgf, Hydroxyurea, Ifna or Ifnb family members, Ifng, Igf1, IL1, IL2, IL4, IL6, IL12, Insulin, ionizing radiation, Lipopolysaccharide (Lps), Neocarzinostatin (NCS), Ngf, Pdgf, Phorbal 12-myristate 13-acetate (PMA), PolyIC, Serum, Sorbitol, Tgfb1, Various TLR9 ligands, Tnf, UV radiation, and Vegf.
In addition we have combined all the maps into one map with all the stimuli (allDish) and one with no stimuli (NoStimDish). The 'no stim' dish has all of the receptors for the ligands, but as the name suggests, no ligands. Effects caused by more than one ligand can be viewed by creating a new dish using the edit command in the "Select Dish" menu. The 'no stim' dish is a convenient starting point (in the dish editor, ask Maude). An example of dish construction is provided in the SmallKB guide. The network displayed by PLA for a given dish is the network of reactions that have been shown to occur in response to the stimulus.
The experimental evidence for each rule is supplied in datum form. Each datum represents a result from a experiment published in a refereed journal. In some cases, the evidence is weak or incomplete or simply based on correlations. Some attempt has been made to point out the weak spots but the bulk of the effort has gone into collecting and presenting the data in such a way that the biologist can decide for himself. Datums are written in a shorthand designed to be both readable by biologists and parsible by a computer. A manual containing the abbreviations and conventions used by the shorthand can be found in the curation notebook . Information about unfamiliar protein names can be found by clicking on the protein representation in the PLA viewer.
The Dishes
Cell biologists spend a lot of time throwing things (stimuli) at cells in culture and studying the results. The size of (number of rules and datums) each dish depends on the available literature and the inclination of the curator to find it.
• Peptide ligands with specific receptors
In some cases, the stimulus is a peptide ligand that binds specifically to one or more receptors. The results depends on which receptors are present on the cell and in a state to bind the ligand. The dishes that use ligands as stimuli are:
|
Dish |
Ligand(s) |
Receptor(s) |
# of Rules |
# of Datums |
|
Csf1Dish |
Csf1 |
Csf1R |
29 |
106 |
|
EgfDish |
Egf |
EgfR |
129 |
2281 |
|
HgfDish |
Hgf |
HgfR |
48 |
329 |
|
IfnabDish |
Ifna family, Ifnb1 |
IfnaR1, IfnaR2 |
14 |
161 |
|
IfngDish |
Ifng |
IfngR1, IfngR2 |
13 |
194 |
|
Igf1Dish |
Igf1 |
Igf1R |
16 |
133 |
|
IL1Dish |
IL1a, IL1b |
IL1R1, IL1Rap |
67 |
1288 |
|
IL2Dish |
IL2 |
IL2Ra, IL4Rb, IL2Rg |
49 |
772 |
|
IL4Dish |
IL4 |
IL4R, IL2Rg |
30 |
281 |
|
IL6Dish |
IL6 |
IL6R, Gp130 |
23 |
282 |
|
IL12Dish |
IL12 |
IL12Rb1, IL12Rb2 |
15 |
313 |
|
InsDish |
Ins |
InsR |
39 |
732 |
|
NgfDish |
Ngf |
NgfR, Trka |
46 |
301 |
|
PdgfDish |
Pdgfa, Pdgfb |
PdgfR |
25 |
203 |
|
Tgfb1Dish |
Tgfb1 |
TgfbR1, TgfbR2 |
57 |
968 |
|
TnfDish |
Tnf |
TnfR1 |
87 |
1915 |
|
VegfDish |
Vegfa |
VegfR2 |
11 |
55 |
• Surface protein ligands with specific receptors
Cd40Lg is a protein that is expressed on the surface of activated T-cells that binds to Cd40 on the surface of antigen presenting B-cells. To avoid confounding factors that arise from using one cell to activate another, the response to Cd40Lg is usually studied by treating cells with an antibody to Cd40.
|
Dish |
Stimulus |
Receptor |
# of Rules |
# of Datums |
|
Cd40Dish |
anti-Cd40 |
Cd40 |
35 |
264 |
• Multiple stimuli with multiple receptors
When studying signal transduction it is usually not helpful to perform experiments in which unknown mixtures of ligands are added to cells. One exception is the addition of serum to cells that have been made quiescent by removal of serum. The addition of serum to cells in G0 phase causes them to re-enter the cell cycle, make new protein, replicate DNA, and divide. This provdes researchers with information about the changes in proteins during the cell cycle regardless of the actual stimuli that caused them.
|
Dish |
Stimulus |
Receptor(s) |
# of Rules |
# of Datums |
|
SerumDish |
Serum |
many |
29 |
303 |
• Stimuli that turn on Pattern Recognition Receptors
The pattern recognition receptors (PRRs) are used by cells to recognize pathogen-associated molecular patterns such as bacterial carbohydrates, nucleic acids, peptides, and lipids. Certain compounds have been found that mimic bacterial products so that the effects of PRR binding can be studied.
|
Dish |
Stimulus |
Receptor(s) |
# of Rules |
# of Datums |
|
LpsDish |
Lps |
TLR2, TLR4 |
98 |
1072 |
|
PolyICDish |
PolyIC |
TLR3 |
25 |
270 |
|
TLR9Dish |
TLR9 Ligands |
TLR9 |
19 |
110 |
• Chemical Stimuli
Researchers often use chemicals to stimulate intracellular events in the absence of receptor binding. Anisomycin and sorbitol cause phosphorylation and activation of the stress responsive Mapks of the Jnk and P38 families. PMA (phorbol 12-myristate 13-acetate) is very similar to diacylglycerol, the natural activator of classic PKC isoforms. Adriamycin is considered to be a mimetic for DNA damage, genotoxic stress that activates Tp53. Bleomycin induces DNA strand breaks and is commonly used a radiomimetic to study the acetylation of ATM and activation of Chek2. Etoposide induces breaks in double stranded DNA and prevents repair by topoisomerase II binding. Hydroxyurea has the capacity to generate DSBs only in replicating cells so it is useful in the study of cross talk between Atm and Atr. NCS (Neocarzinostatin) intercalates into DNA and induces double-strand breaks which makes it another compound that is used to mimic ionizing radiation in signal transduction studies.
|
Dish |
Stimulus |
Targets(s)/Effect(s) |
# of Rules |
# of Datums |
|
AdriamycinDish |
Adriamycin |
Tp53 |
8 |
65 |
|
AnisoDish |
Anisomycin |
Jnks and P38s |
27 |
191 |
|
BleomycinDish |
Bleomycin |
DNA breaks |
8 |
39 |
|
EtoposideDish |
Etoposide |
DS-DNA strand breaks |
15 |
159 |
|
HydroxyureaDish |
Hydroxyurea |
DS-DNA strand breaks |
7 |
32 |
|
NCSDish |
NCS |
DS-DNA strand breaks |
10 |
59 |
|
PMADish |
PMA |
Pkcs |
33 |
309 |
|
SorbitolDish |
Sorbitol |
Jnks and P38s |
11 |
66 |
• Other stresses
Two major stresses that cause DNA damage are ionizing radiation (IR) and ultraviolet light (UV). IR causes double strand DNA breaks and the activation of Atm and Prkdc whereas UV causes single strand DNA damage and the activation of Atr. UV also leads to the activation of Jnks and P38s and phosphorylation of Tp53. Thus, IR and UV are valuable tools for studying the activation of cell cycle checkpoints.
|
Dish |
Stimulus |
Targets(s)/Effect(s) |
# of Rules |
# of Datums |
|
UVDish |
Ultra Violet Light |
dsDNA |
45 |
490 |
|
IRadDish |
Ionizing Radiation |
ssDNA, Jnks and P38s |
29 |
910 |
The purpose of Pathway Logic models such as STM7 is to:
- help biologists keep track of published experimental results
- assist in hypothesis making
- arrange (and rearrange) the curated data into conclusions (graphically)
- locate holes in experimental evidence things that have been accepted as truth for so long that they have become the truth without any evidence
- integrate pathway components into a common framework to see connections between proteins in various "pathways" not discussed in the experimental or review literature
- provide accurate maps suitable for computational analysis
This demo assumes that the user is already familiar with the use of PLA as described in the SmallKB guide and the PLA reference manual .
In this guided tour, double click on a figure to see an enlarged version. Single click to restore the figure to its original size.
Guided Tour: Revisiting the Egf Pathway using PLA and the STM7 KB.
Introduction
In this guided tour we focus on response to Egf stimulation. This version of the Egf Network looks different from others because it is based on experimental data only. An Egf rule is derived from changes observed in response to Egf binding to the EgfR. If assumptions about protein interactions based on over-expression or test-tube reactions or similarity with other tyrosine kinase receptors are used, notes are included in the evidence files. The change in state of the components of the pathway are recorded as Pathway Logic rules which are then assembled into Petri net format by PLA. The resulting graph is loaded into PLA for viewing and analysis. A link to the evidence supporting each rule is provided in the viewer. Once a Petri net is assembled, PLA can be used to find Paths through the network to a chosen goal. The path we are going to discuss in this demo is the path between Egf binding to the EgfR and activation of Erk1 and Erk2 (abstracted as Erks as most of the existing evidence does not distinguish between these two members of the family). This path is traditionally considered to be:
References:
- Biocarta
- The Cancer Cell Map at Memorial Sloan-Kettering Cancer Center.
- Kegg
- Reactome
The STM7 Demo
If you haven't already started the STM7 online demo, you can do so by clicking here and following the instructions.
When PLA starts up, a knowledge base manager window (title: PLA KBManager) appears in the upper left corner of your screen. The KBManager displays a list of available knowledge bases (rule sets). "RKB" is selected by default and will be highlighted in blue.
Start by selecting a predefined dish: press the "Select Dish" button on the right side of the KBManager window. This produces a menu with two options: "Edit" and "PreDefined". Select (click on) "PreDefined". This produces a (sub)menu with a list of pre-defined dishes. Select "EgfDish" (by clicking on the corresponding menu item).
Shortly a PLA viewer window with tab titled graph2:EgfDish will appear on your screen displaying the Egf Network (the network of reactions reachable starting from the occurrences in EgfDish). [Note that the graph numbers in titles may differ if you have created graphs or nets in addition to those discussed in the demo. This is fine.]
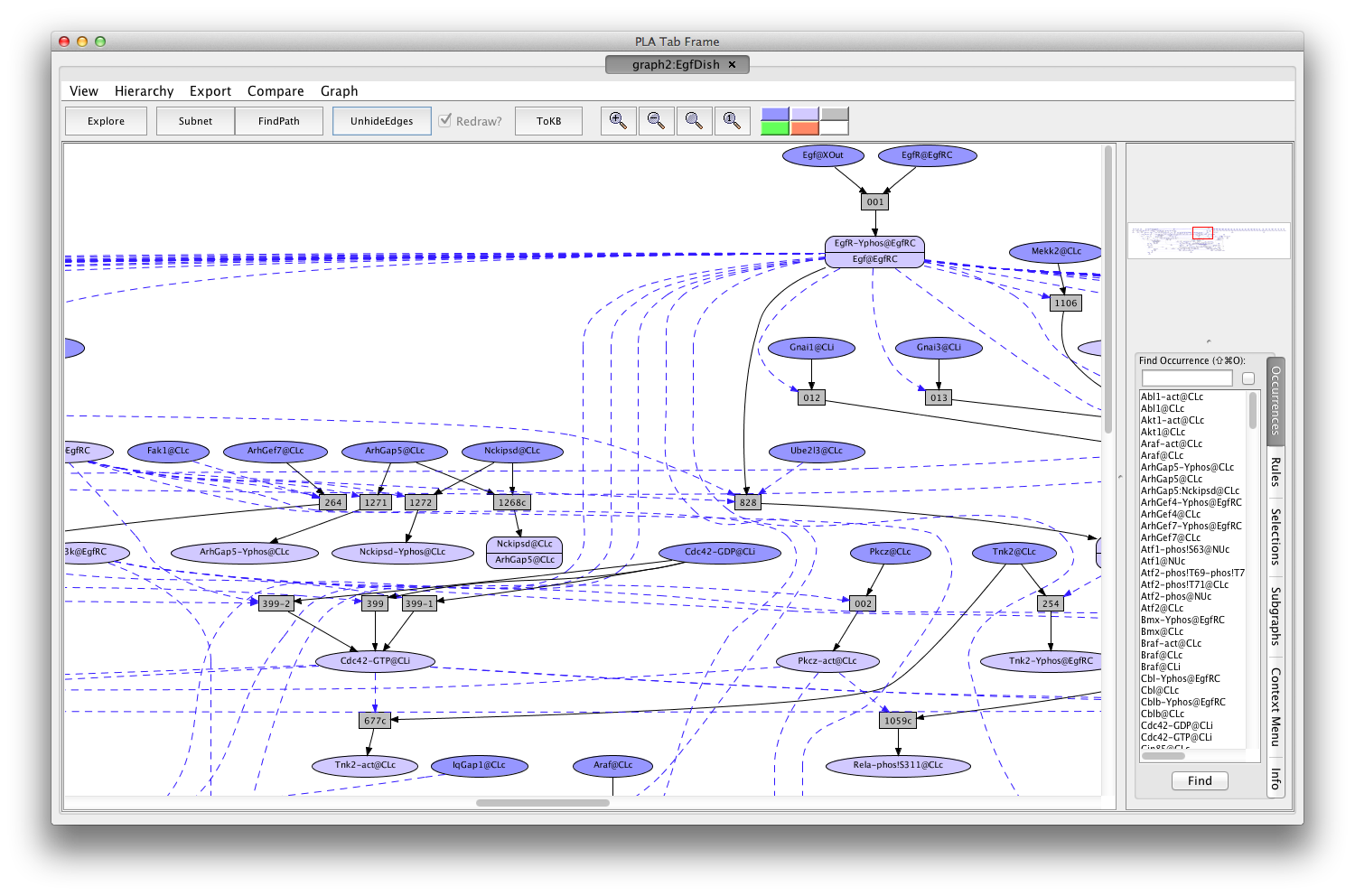
Screen shot of the Egf dish graph
A network of reactions is displayed as a graph with two kinds of nodes. Ovals represent occurrences—proteins or chemicals in a specific state and location. For example the oval labeled Cdc42-GDP@CLi represents the protein Cdc42 bound to GDP and located on the inside (cytoplasmic side) of the cell membrane (@CLi).
We use the following abbreviations.
Locations
Loc |
Description |
Loc |
Description |
XOut |
outside the cell, the medium or supernatant |
CLm |
in/across the cell membrane |
CLi |
attached to the inside of the cell membrane |
CLc |
in the cytoplasm |
NUc |
in the nucleus |
Sig |
a way to display comments or cellular phenotypes in a network |
Receptor Complexes
Receptor |
Description |
Receptor |
Description |
Cd40C |
forms in response to Cd40Lg binding to Cd40 |
Csf1RC |
forms in response to Csf1 binding to Csf1R |
EgfRC |
forms in response to Egf binding to EgfR |
GP130C |
forms in response to IL6 binding to Gp130 and IL6R |
HgfRC |
forms in response to Hgf binding to HgfR |
IfnaRC |
forms in response to members of the Ifna family or Ifnb1 binding to IfnaR2 and IfnaR1 |
IfngRC |
forms in response to Ifng binding to IfngR1, Jak1, IfngR2, and Jak2 |
Igf1RC |
forms in response to Igf1 binding to Igf1R |
IL1R1C |
forms in response to IL1 binding to IL1R1 and IL2Rap |
IL2RC |
forms in response to IL2 binding to IL2Rb, IL2Ra, and IL2Rg |
IL4RC |
forms in response to IL4 binding to IL4R IL2Rg |
IL12Rb1C |
A preassembled complex consisting of IL12Rb1 and Tyk2 |
IL12Rb2C |
A preassembled complex consisting of IL12Rb2 and Jak2 |
IL12RC |
A complex that forms in response to IL12 binding to IL12Rb1, Tyk2, IL12Rb2, and Jak2 |
IL22RC |
forms in response to IL22 binding to IL22Ra1 and IL10Rb |
InsRC |
forms in response to Ins binding to InsR |
NgfRC |
forms in response to Ngf binding to NgfR |
PdgfRC |
forms in response to Pdgf binding to PdgfR |
Tgfb1RC |
forms in response to Tgfb1 binding to TgfbR1 and TgfbR2 |
TLR2C |
forms in response to TLR2 ligands binding to TLR2 |
TLR3C |
forms in response to TLR3 ligands binding to TLR3 |
TLR4C |
forms in response to TLR4 ligands binding to TLR4 |
TLR9C |
forms in response to TLR9 ligands binding to TLR9 |
TnfR1C |
forms in response to Tnf binding to TnfR1 |
TrkaC |
forms in response to Ngf binding to Trka |
VegfR2C |
forms in response to Vegfa binding to VegfR2 |
Non Receptor Complexes
Non Receptor |
Description |
Non Receptor |
Description |
RNAC |
forms around the capped mRNA complex |
CHR |
forms around Chromatin |
Modifications
Modification |
Description |
Modification |
Description |
acetyl!site |
acetylated on a specific site |
act |
activated |
degraded |
degraded |
dimer |
dimerized |
GDP |
bound to GDP |
GTP |
bound to GTP |
K48ubiq |
covalently bound to ubiquitin polymerized via K48 linkages |
K63ubiq |
covalently bound to ubiquitin polymerized via K63 linkages |
p50 |
a 50kD cleavage product |
phos |
phosphorylated |
phos!site |
phosphorylated at a specific site |
sumo |
sumoylated |
ubiq |
ubiquitinated |
Yphos |
phosphorylated on tyrosine |
off |
not transcribing mRNA |
on |
transcribing mRNA |
Darker colored ovals represent occurrences in the initial state (the selected dish when the network is generated by choosing a dish). Lighter colored ovals represent potential states/locations of these components.
Rectangles represent rules. The label in a rectangle is its (abbreviated) identifier in the knowledge base. Solid arrows from an occurrence to a rule indicate that the occurrence is a reactant (rule input). Solid arrows from a rule to an occurrence indicate that the occurrence is a product (rule output). Dashed arrows from an occurrence to a rule indicate that the occurrence is a modifier/enzyme/catalyst—it is necessary for the reaction to take place but is not changed by the reaction. The graph has been simplified by checking the Redraw? box followed by clicking on the 'HideEdges' button in the tool bar. This operation removes redundant edges. Meaning: if there are edges connecting A → B → C and A → C, then the edge leading from A to C will be removed, as it is logically implied by the other two edges. In the sequel all graphs will have been so simplified.
Subnets and Pathnets
The network shown in the graph2:EgfDish includes all the reactions that fire in response to Egf. Suppose we are only interested in those reactions leading to the activation of Erks. This is accomplished by finding the occurrence that contains activated Erks, marking it as a goal, and asking for the relevant subnet:
- Select Occurrences tab (in the lower right panel).
- Type Erks in the Find Occurrence window.
- Press on the Tab key until "Erks-act-phos(TEY)@CLc" is highlighted in the list below.
- Click on the Find button. This will locate and highlight the requested occurrence node in the main view window.
- Click on this node to open the context tab in the lower right panel.
- Click on the "make occ a goal" button in the context window. The node will turn green.
- Press the button labeled Subnet in the tool bar. Shortly a new tab will appear displaying a graph representing a subnet containing all the paths leading to activation of the Erks. (graph4:S(graph2)).
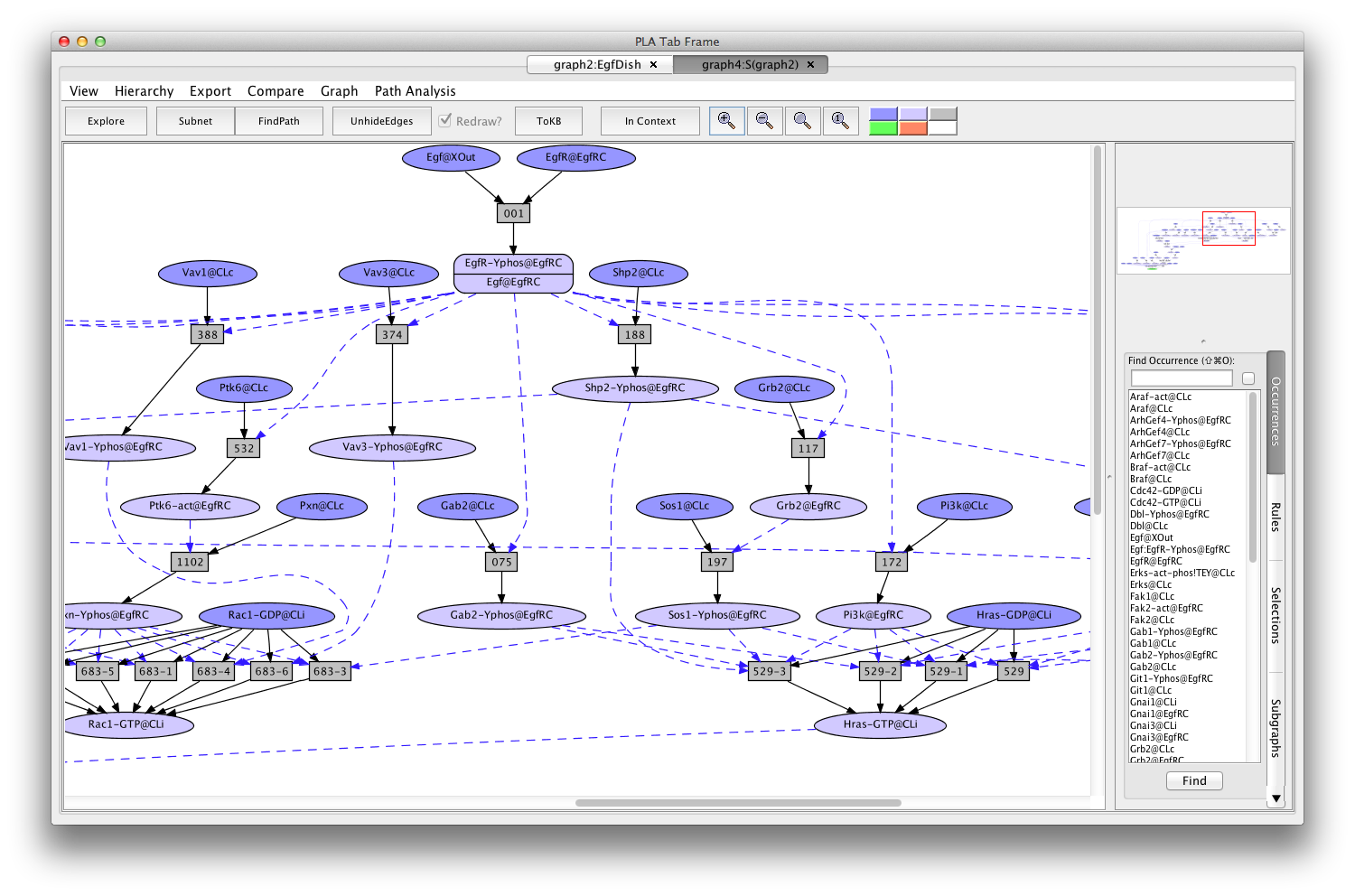
Screen shot of the subnet relevant to activation of Erks
Graph4 is still a network in that it contains more than one route to get to the Erks-act-EgfR goal. Clicking on the FindPath button in the tool bar will find one path (usually the shortest).
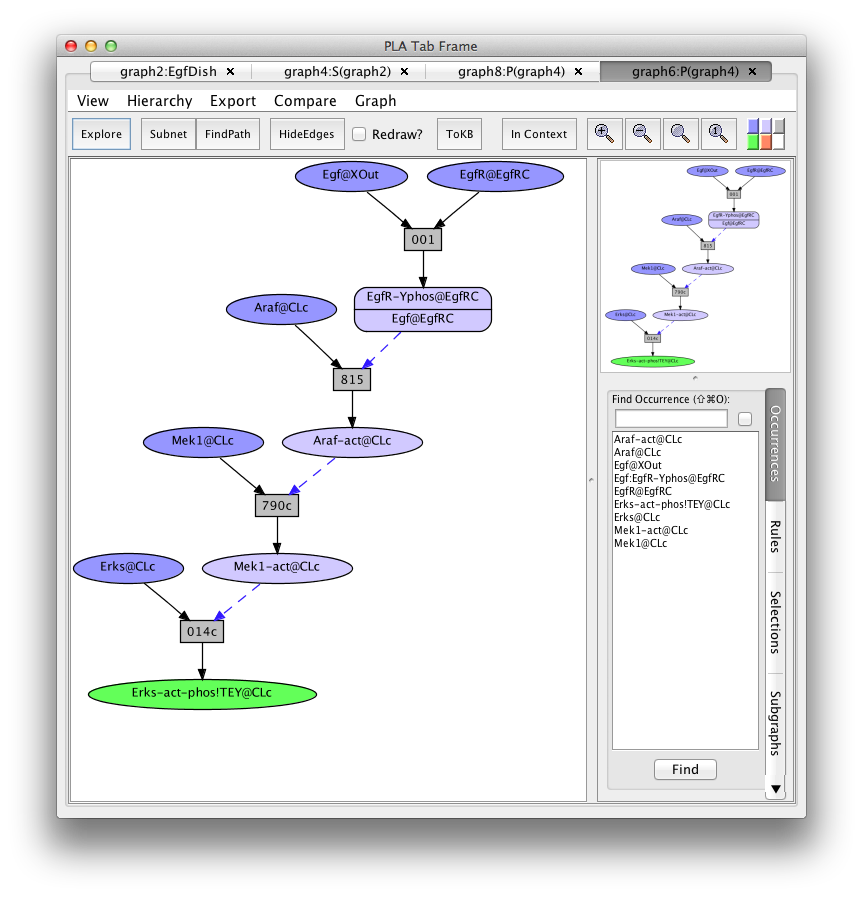
Screen shot of a pathway activating of Erks
Graph6 is called a pathnet because the set of rule nodes represent a particular flow of signal from initial state to goals (called an execution or computation).
This path is particularly simple because it uses "common rules" (indicated by "c" at the end of the rule number) that lack detailed biological context information. To find another path, go back to graph4, click on rule 014c, click on the "hide rule" button in the context tab on the right, and click on "FindPath" again (graph8:P(graph4)).
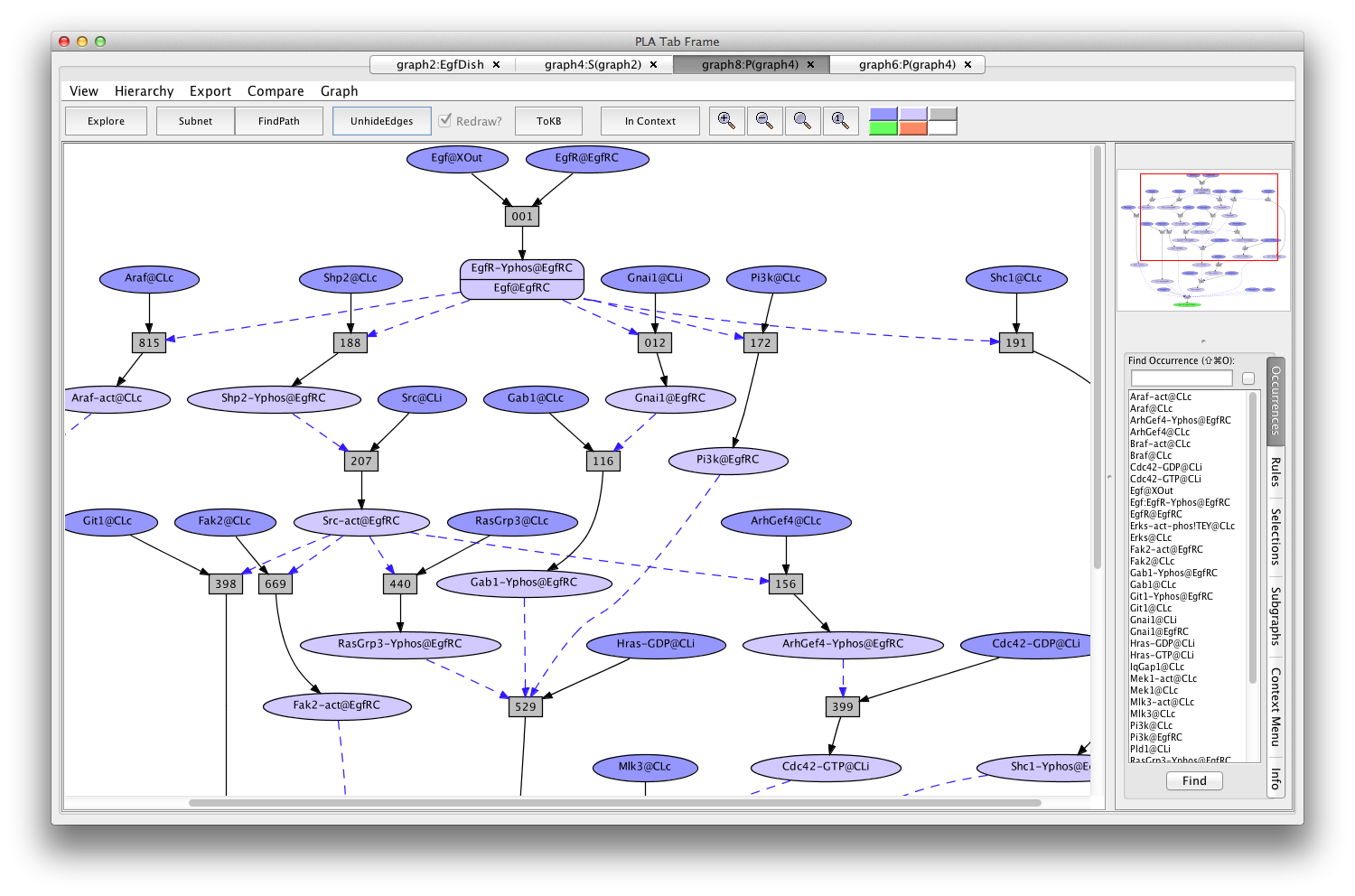
Another Erks pathway
If the Pathnet in graph8 is compared to the Subnet in graph4 (choose (graph4:S(graph2)) from the Compare menu), nodes of the path will be pink and alternative nodes will be highlighted in teal (graph11:C(graph8,graph4) ).
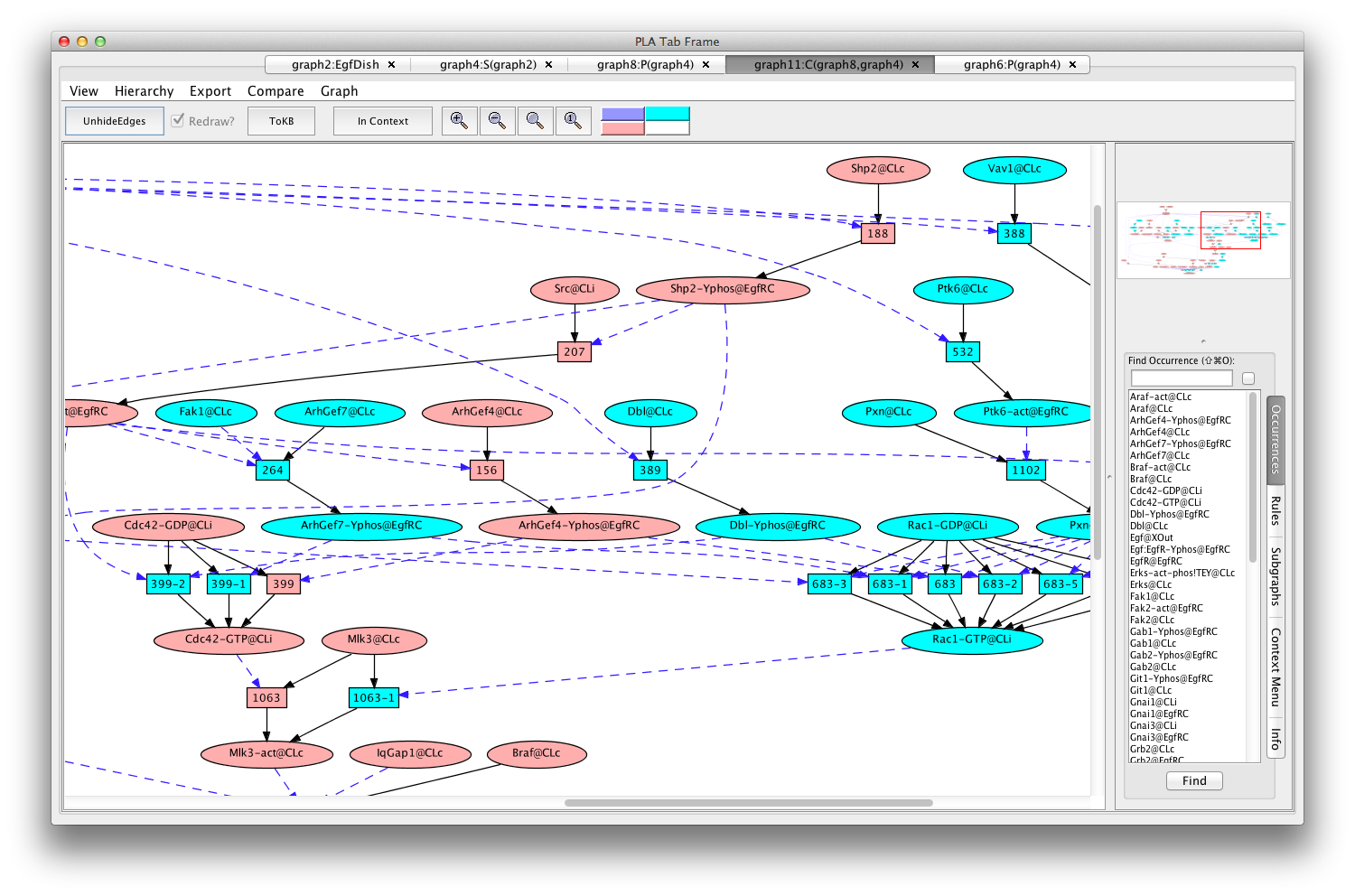
The Erks pathway in the context of the subnet (edges hidden)
Comparison of the subnet with the pathnet shows that there is a choice of two major routes to the activation of Mlk3. Either Cdc42 or Rac1 can activate Mlk3. Cdc42 and Rac1 themselves are activated by different GEFs (guanine nucleotide exchange factors).
A subnet using Rac1 instead of Cdc42 can be generated by returning to the graph4 (the Erks subnet of EgfDish), marking Cdc42-GDP-CLi as an avoid (keeping 014c hidden) and pressing the FindPath button again (graph13:P(graph4)).
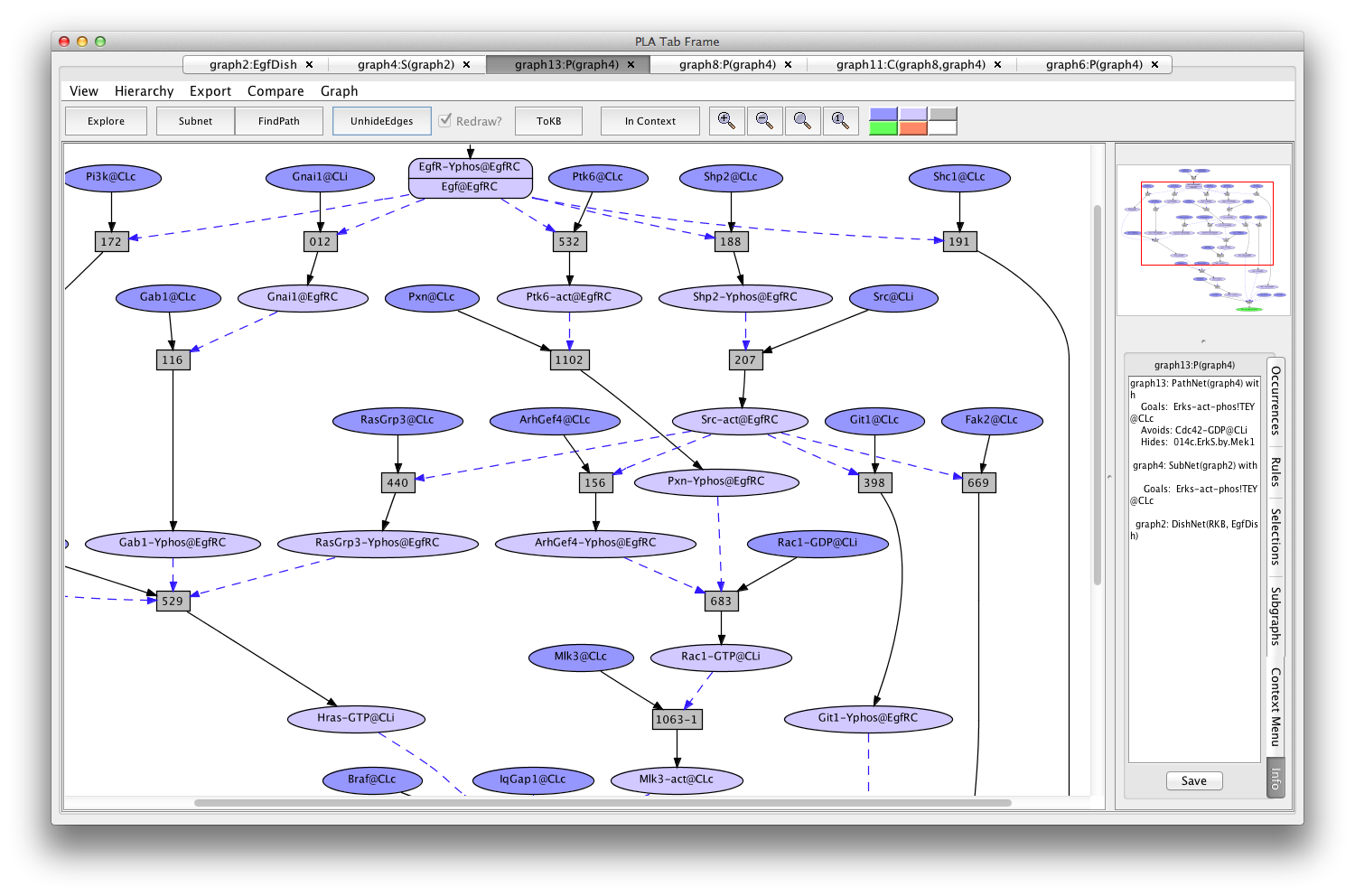
Screen shot of a pathway activating Erks avoiding Cdc42
You can compare the two pathways by selecting the "Compare" menu from the menu bar, and selecting the original pathnet (the only one available). The pink shows the shared elements of the pathways, while the blue and teal indicate elements unique to the different pathways.
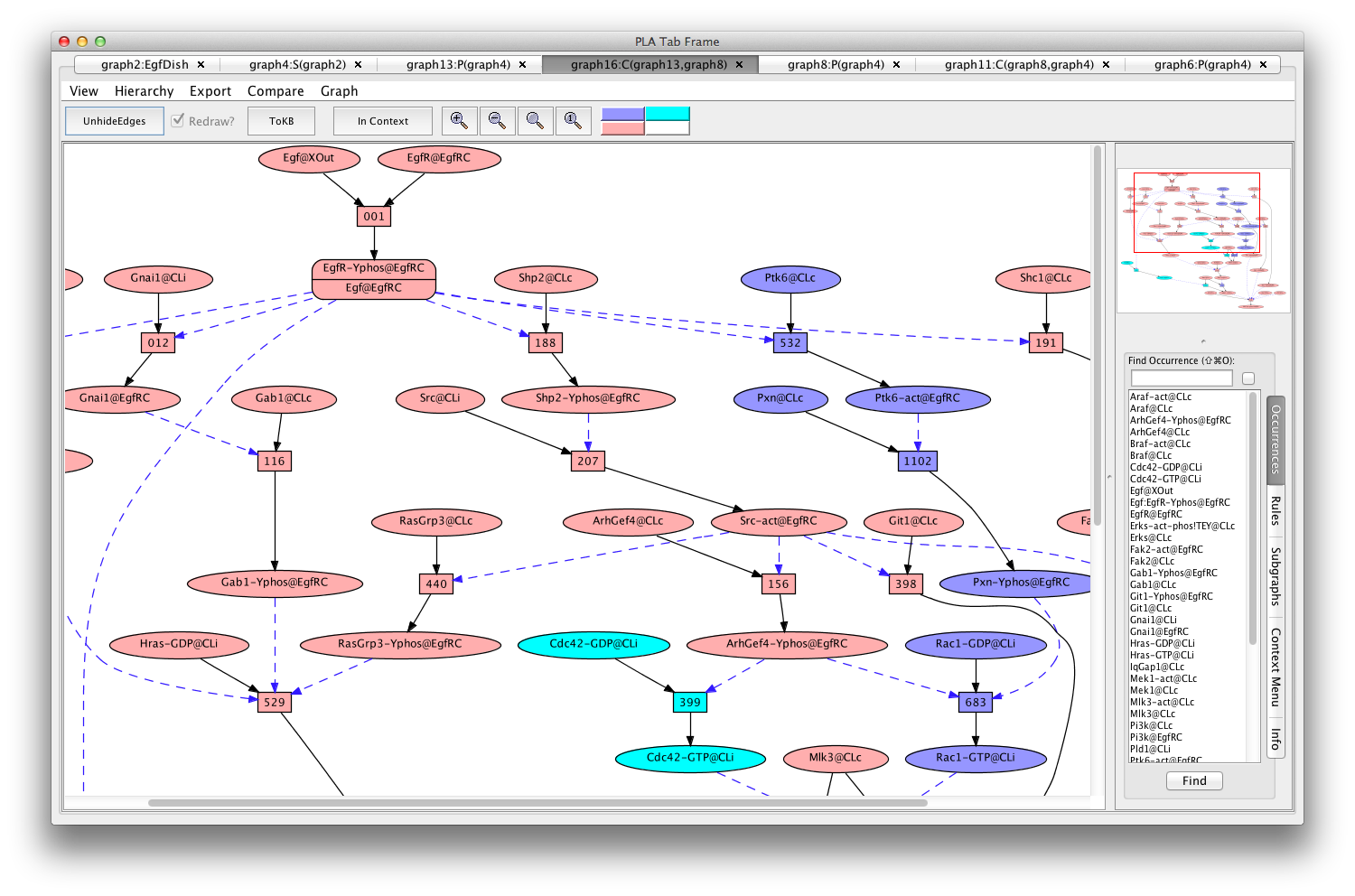
Comparing pathways activating Erks using and avoiding Cdc42
Rules
The little gray rectangular nodes represent rules. A rule describes the transition of the state of the proteins in a cell to another state. Only the proteins involved in the transition are mentioned in the rule. Consider rule 196 at the bottom of the Pathnet (graph8). (You can see this rule in its own window by clicking on the rule node and then clicking on the "exploreRule" button in the resulting information window. The rule is now displayed in an explorer window and the node colors indicate whether there are any connections in the parent network. Place the mouse over the color key in the upper right for details.) The rule says that if the following occurrences are present:
IqGap1@CLc, Braf-act@CLc, Mlk3-act@CLc, Pi3k@EgfRC, Fak2-act@EgfRC, Hras-GTP@CLi, Erks@CLc, Pld1@CLi, Shoc2@CLc, Gab1-Yphos@EgfRC, Git1-Yphos-EgfRC, Shp2-Yphos@EgfRC, Mek1-act@CLc, Shc1-Yphos@EgfRC, Shc1-Yphos@EgfRC, Src1-act@EgfRC and the Egf:Egfr complexthen the Erks will become TEY phosphorylated and activated.
Evidence.
Where did all those bizarre requirements come from? Clicking on the rule node will open the information window. Clicking on the Show Evidence button will call up a document containing the experimental evidence that supports (or not) the rule.
The evidence page is written in datum shorthand and will look at first to be uninterpretable. Instructions on how to read the evidence can be found in the curation notebook. For those who find it easier to read each reference and dig out the evidence for themselves, we have provided the PubMed ids and the Figure/Table numbers of the relevant experiments.
Discussion of rule 196.
The evidence page for rule 196 reports on 237 experiments in which Erk1, Erk2, or both were either phosphorylated on their TEY site (T202/Y204 for Erk1, T185/Y187 for Erk2) or activated in an in vitro kinase assay. The peak response occurred around 5-10 min. Forty-nine different cell lines or primaries were represented.
Things that are required
The requirements listed above for the activation of Erks in response to Egf (rule 196) are determined by performing the stimulation in cells lacking proteins due to knock-out at the gene level [KO], knock-down by RNA interference [RNAi], mutation at the gene level [substitution], or expression of a dominant-negative mutant [addition]. Some of the required proteins such as Mek1, Hras, and Shc1 are familiar and mentioned in most of the published Egf Pathway diagrams. Surprisingly, although Raf1 is activated in response to Egf, there is plentiful evidence that Raf1 is NOT required for Erk phosphorylation or activation in response to Egf
Surprisingly, although Raf1 is activated in response to Egf, there is plentiful evidence that Raf1 is NOT required for Erk phosphorylation or activation in response to Egf [11296227, 11296228, 12048245, 17314031, 15258589]
References [17314031, 11296228] also show that Mek1/2 activation in response to Egf does not require Raf1.
So what activates Mek1? Other kinases capable of phosphorylating Meks at the SMANS site (S218/S222 for Mek1, S222,S226 for Mek2) are Braf [17314031], Araf [8621729], Pak2 or Pak3 [17314031], and Mlk3 [12738796].
Could Araf substitute for Raf1? Evidence that Araf is activated in response to Egf is equivocal (click here) and although Araf has been shown to be capable of increasing the kinase of activity of Mek1 in a cell-free system no experiments have be found testing the requirement of Araf for either Mek1-phos/act or Erks-phos/act in response to Egf.
Activated Pak1, Pak2, and Pak3 can phosphorylate Mek1 directly on S298 which leads to Mek1 autophosphorylation on (S218/S222) [17314031 Fig-4a].
Although Pak1 has been shown to be activated in response to Egf, phosphorylation of Meks and Erks at their activation sites in response to Egf was not affected by RNAi to Pak1 and Pak2 [16129686-Fig-3a]. In addition, there are reports that Erks are not activated when cotransfected with active Pak1 [8805275 Fig-5a, 9351825 Fig-6b, 16698790 Fig-6b].
Pak3 can be excluded because it is not activated irt Egf [11259591-Fig-5a].
Although Mlk3 can phosphorylate Mek1 directly [12738796] on an unknown site, its requirement by Braf and Erks does not require its kinase activity [16537381].
Furthermore, over-expression of Mlk3 actually inhibits the activation of Erks in response to Egf [12738796].
This is indicative of a scaffold or adaptor protein rather than a active member of a phosphorylation cascade. Mlk3 is required for activation of Braf [16537381], Mek1 [15258589] and Erks[15258589, 16537381] in response to Egf.
It is unfortunate that the papers that demonstrated that Braf was required for Erk phosphorylation [15199148, 15258589] did not perform one additional experiment to show that Braf is required for Mek1 activation in response to Egf. One of the objectives of Pathway Logic is to make it easier to discover such omissions.
Src and Fak2 are recruited to the EgfR complex and phosphorylated in response to Egf so it is logical to wonder if they are part of the path that activates Erks. Erk activation is only partially inhibited by expression of dominant-negative Src and Fak2. A partial inhibition suggests that there may be something that substitutes for the proteins. That idea is supported by the information that activation of Src in response to Egf requires Rala [10675331] but Erk phosphorylation does not require either Rala [10675331] or RalGds [11283245, 18285454].
The locations
The interactions of some of the required proteins such as Fak2, Shc1, Sos1, Src, and Git1 with phosphorylated EgfR have been extensively studied so that we have information about their change in location and modifications in response to Egf. Others such as Shoc2 and IqGap1 are recent findings and more curation (or experimentation) is needed to determine location and modifications. These proteins have been placed in the cytoplasm without modifications until more information is available.
Interestingly we have been unable to find any experiments in which Hras, Braf, Mek1, or Erks have been shown to coprecipitate (or not) with EgfR after Egf stimulation. Thus the accumulation of these proteins into the EgfR complex are merely a supposition of the curator. The lack of evidence for the location is duly noted at the top of each evidence file.
Things that are missing
A noticeable discrepancy is that Mek2 is not mentioned. Out of 43 experiments in which changes to Meks in response to Egf are recorded, only one of them looks at Mek2 specifically. In the cases where an anti-phospho-Mek antibody was used, the antibody was reported to be specific for only phospho-Mek1 or for both phospho-Mek1 and Mek2. There were no experiments that used an antibody specific for phospho-Mek2. Therefore, only Mek1 is represented in this pathnet.
Conclusions
The network presented here is not intended to be cast in stone. It is a set of hypotheses derived from the curated data that provide a logical explanation of how a signal propagates in response to an Egf stimulus.
This demo provides an illustration of:
- How PLA can be used to help interpret experimental results.
- The ease with which pathways can be assembled, drawn, and navigated.
- How collecting experimental results in the form of datums can aid reasoning and discussion.
- How gaps in experimental knowledge can be discovered in the process of formulating and checking the hypotheses.