Overview of the Mycobacterial Mycolic Acid Biosynthesis Pathway
What is Mycolic Acid?
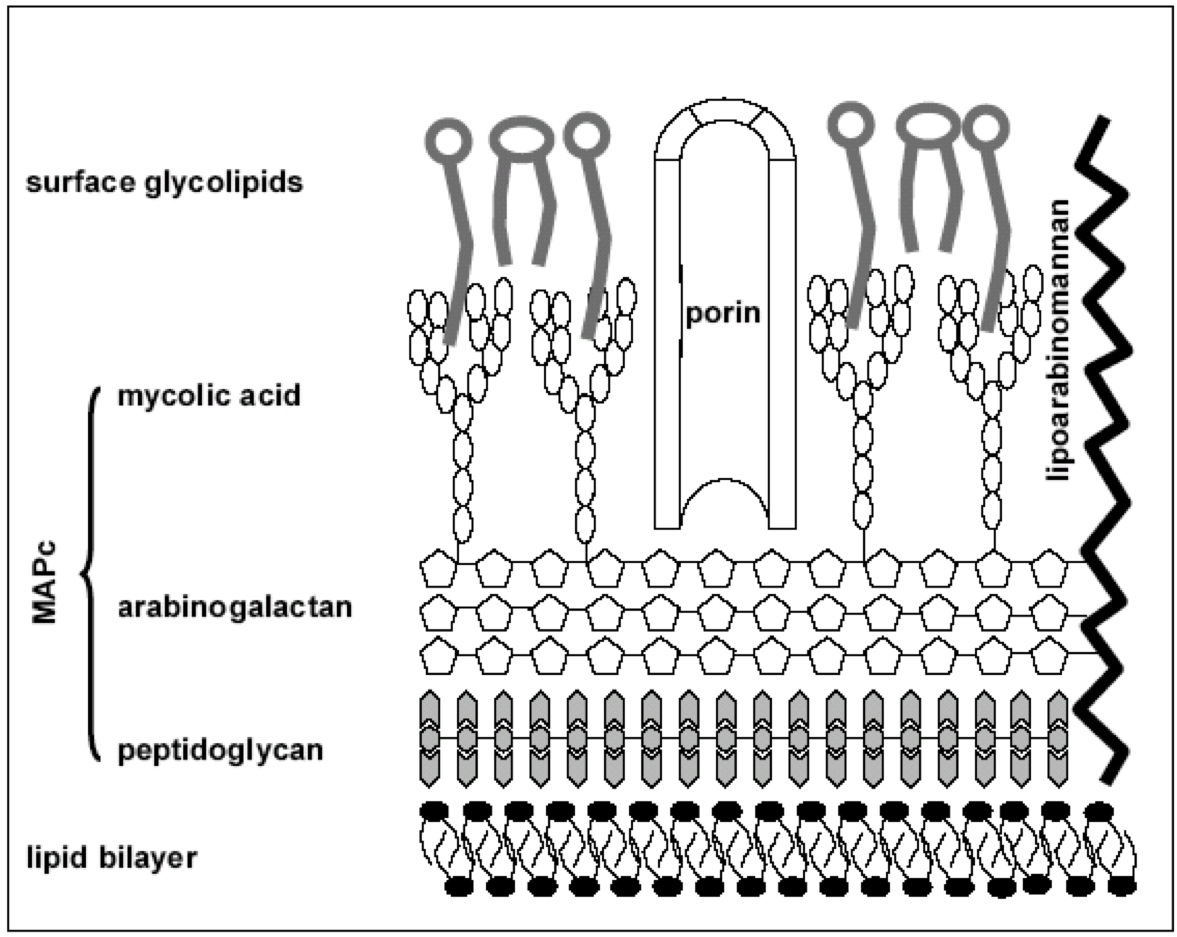
Mycolic acids, a homologous series of C60-C90 long-chain
alpha-alkyl-&beta-hydroxy fatty acids, represent essential components
of the mycobacterial cell wall. They are important for mycobacterial
growth, survival, and pathogenicity. They are found as esters of an
arabinogalactan as well as free lipids in the form of trehalose di
mycolate (TDM). Arabinogalactan-mycolate is covalently linked to the
cell wall peptidoglycan via a phosphodiester bond located on the inner
leaflet of the outer membrane. Both arabinogalactan-mycolate and TDM
provide a protective thick cell wall and protect the tubercle bacillus
from antibiotics and host's immune system. TDM also inhibits
phago-lysosome fusion and is often considered to be an indicator of
virulent strains.
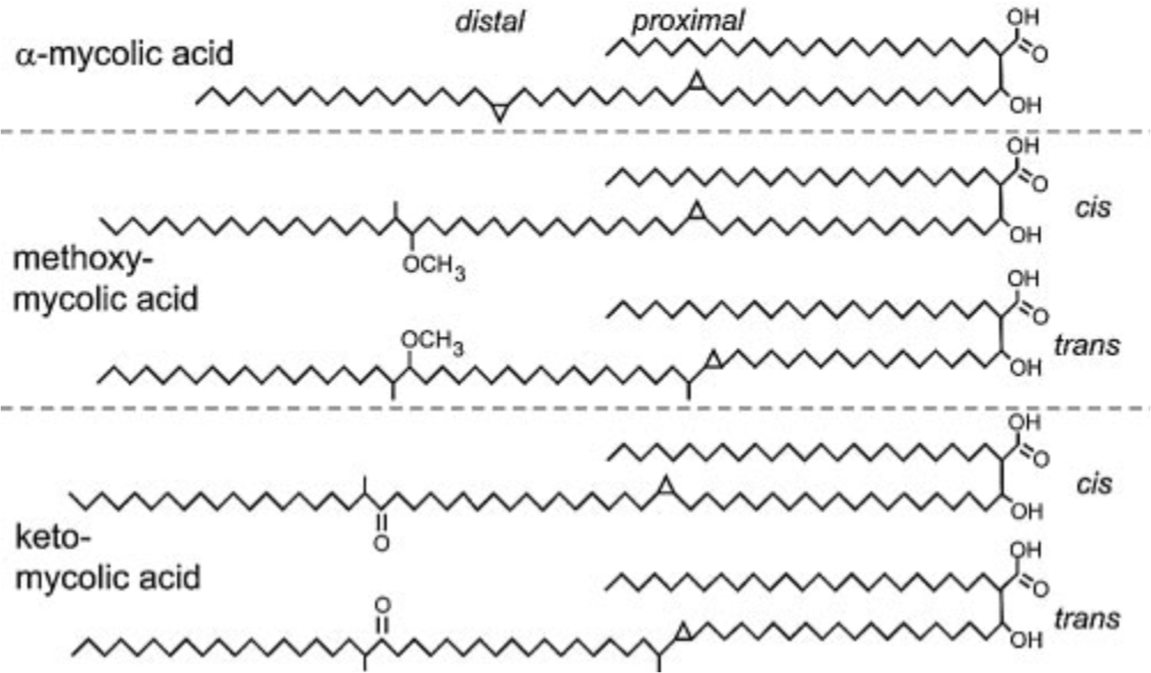
Three distinct structural classes of mycolic acids namely alpha- (more
than 70 percent), methoxy- and keto-mycolic acids (10-15 percent) are
found in this bacillus. The alpha-mycolic acid is a
cis,cis-dicyclopropyl fatty acid. Both methoxy- and keto-mycolic acids
have either cis- or trans-cyclopropane rings. Cyclopropane rings in
mycolic acids protect the bacillus from oxidative stress
[
15653820,
17555433,
15353567
].
Mycolic acid pathway as TB drug targets
Several front-line drugs used for treating tuberculosis inhibit
mycolic acid synthesis. Understanding the pathway of mycolate
biosynthesis therefore helps understand the underlying molecular
mechanisms of the disease tuberculosis as well as the identification
of new anti-tuberculosis drug targets. InhA (EC 1.3.1.9,
enoyl-[acyl-carrier-protein] reductase), involved in mycolic acid
synthesis, is a target for front-line anti-tubercular drugs, such as
isoniazid and ethionamide
[
16261191].
Enzymes needed for biosynthesis of mycolic acids, such as methyl
transferase (PcaA), beta-ketoacyl-acyl carrier protein synthase (KasAB
and FabH), acyl-AMP ligase (Fad32) and polyketide synthase (Psk13) are
promising drug targets for new anti-TB agents
[
20477209].
Critical steps of the pathway
Mycolates are synthesized by at least two discrete elongation systems
in Mycobacteria - the type I and type II fatty acid synthases (FAS-I
and FAS-II respectively). The eukaryotic-like FAS-I is a single
polypeptide with multiple catalytic activities (encoded by the fas
gene, Rv2524c) that catalyses the de novo synthesis of fatty acids
from acetyl-CoA. The domains of M. tuberculosis FAS are organized in
the following order: acyltransferase, enoyl reductase, dehydratase,
malonyl/palmitoyl transferase, acyl carrier protein, beta-ketoacyl
reductase, and -ketoacyl synthase. FAS-I exhibits a bimodal product
distribution: C20 and C26 acyl CoAs. These form the substrates for the
FAS-II reaction cycle and the polyketide synthase enzyme
respectively. Beta-ketoacyl-ACP synthase III forms a pivotal link
between FAS-I and FAS-II. FAS II, similar to that in other bacteria,
consists of dissociable enzyme components, which act on a substrate
bound to an acyl-carrier protein (ACP). Four condensases participate
in initiation (FabH), elongation (KasA and KasB) and termination
(Pks13) steps, leading to full-length mycolates. Synthesis of mycolic
acids in the cell is followed by extensive postsynthetic modifications
and unsaturation reactions. Condensation of the fully functionalized
and preformed meromycolate chain with a 26-carbon alpha-branch
generates full-length mycolic acids that must be transported to their
final location for attachment to the cell-wall arabinogalactan. Three
well-known immunogenic proteins of the antigen 85 complex mediate the
transfer and subsequent transesterification. The basic reactions in
FAS-I and FAS-II are a repetition of a cycle of four reactions, each
cycle culminating in the extension of the alkyl chain by a two-carbon
unit
[
15653820,
15353567].
A mycobacterial mycolic acid biosynthesis model has been developed in pathway logic.
A guided tour of
this model can be found
here .
You can browse and analyse the Pathway Logic mycolic acid biosynthesis model
using the Pathway Logic Assistant
client
. Just click on the link and follow instructions.
The guide includes suggestions for subnets to explore
and instructions for creating them using the Pathway Logic Assistant
viewer.