PLA Datums
Table of Contents
Introduction
The Pathway Logic (PL) STM model (available here) is a network of protein interactions and modifications which are used by the cell to transmit signals from its environment to the nucleus. The purpose of the model is to display the various ways the reactions can be connected in order to study the effects of removal/inhibition/mutation of proteins on downstream events. The reactions are derived from published experimental evidence and the problem of how to store and search the evidence became a project in itself. We eventually developed a "shorthand" language made up of modules that can be read by biologists, traced back to their source, and have enough structure to be interrogated computationally. We call those modules "datums".
The PL Datum Knowledge Base (DKB) has grown to over 77,000 datums related to the state of mammalian cells and their response to a variety of stimuli. In order to share the KB with the scientific community, we have made a web based interface that can be used to search for specific experimental results and answer questions related to protein interactions and modifications.
This document provides an introduction to datums and describes how to search the DKB using the web based interface (http://stella.csl.sri.com).
How to Read a Datum
A datum is a summary of an experimental finding curated from a published data paper (normally a paper indexed by Pubmed). Each datum represents one biological assay.
The Two Types of Datum
There are two main types of datum, state and change datums, corresponding to two basic types of biological experiments.
State datums describe the state of something in a defined system. An example of a state experiment is the demonstration of the existence of a protein in a particular cell line using a Western Blot. Protein interaction data is another example of the use of a state datum. If one protein can be co-precipitated by another protein from the same cell, they are considered to interact. State data is used by Pathway Logic to create initial system states -- a list of components, modifications, and locations that are then subjected to rules that define changes in the state in response to various stimuli.
The rules are derived from change datums which summarize the change in the state of something resulting from the addition of a stimulus to cells. An experiment in which the phosphorylation of the Egf receptor is increased after addition of Egf to a cell for 5 minutes is an example of a change datum.
Datum Structure
A datum can be parsed by a computer because it uses a structured syntax and a controlled vocabulary. It consists of at least three lines. The first line contains the protein(s) that are observed (Subject), the assay (Assay), the stimulus (Treatment), and a result (Change). The second line contains the cells and culture conditions that were used in the assay (Environment). The last line (Source) gives the PubMed ID (PMID) of the source article and the number of the figure containing the experimental result. There may be additional lines, called "extras", describing variants on experimental conditions.
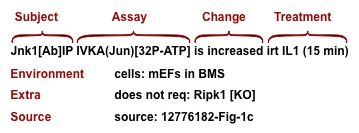
The Major Elements
Subject: The subject (mostly proteins, some genes) is what the experiment is about. Protein subjects carry attributes that provide additional information including the origin (for example expressed or endogenous), how it is identified (by antibody to the protein, by antibody to the tag on an expressed protein, or by some other sort of label), and whether or not it is an immunoprecipitate.
Assay: The assay is the experimental method. For example Prot-exp[WB] denotes an assay in which total protein expression in a cell is detected by Western Blot, and Surface-exp[FACS] denotes an assay in which protein expression on the surface of a cell is detected by Flow Cytometry. Some assays have assay specific attributes such as hook for binding assay, and substrate for activity assay. A catalog of assays and their attributes can be found in the Assays section of the Datum Dictionary.
Treatment: The treatment can be addition of a ligand (e.g., a peptide or chemical), expression of a protein, or a stress (such as UV light).
Result: In a state type datum the result is either detectable or undetectable. In a change type datum the result has two parts -- the change (increased, decreased, unchanged, or detectable-but unchanged) and the time(s) after addition of a stimulus that the result is observed. If multiple time points are used they are treated as a separate datum element and each time has an associate qualitative level of change indicated by 0 or more "+"s following the time.
Environment: The environment of a datum describes the cell and medium in which the experiment is performed. Cell types can be cell lines or primary cells. Additional information can include exogenous proteins that have been over-expressed in the cells or endogenous proteins that have been removed by knockout or RNA interference technologies. The medium is described with minimal detail using defined abbreviations such as `BMS' for basal medium with serum.
Extras: Extras are use to record the effect of an alteration to some component of an experiment. The most common use of an extra is the observation of the effect of a small molecule on a result. A typical example is the inhibition of the phosphorylation of Akt1 in response to Egf by Wortmannin. Other uses include the effect of depletion of endogenous protein by inhibitory RNAs or the replacement of an expressed protein by a mutated form.
Source: The source is usually a PubMed ID together with a Figure or Table number. It could also be an unpublished laboratory result.
The Minor Elements
In addition to the major elements, datums contain clues about experimental details that are important to the interpretation of the results. The example displayed below says that the coprecipitation of expressed Mek1 by endogenous Erk2 is increased when constitutively active ("CA") Rac1 is also expressed.

The information that Mek1 is expressed is supplied by the prefix "x" before the name of the protein. The handle "tAb" says that the antibody used to detect Mek1 is against a tag on the expressed protein rather than the protein itself. Endogenous proteins do not have prefixes. The assay is named "copptby" and "WB" (western blot) is the detection method. The detail that expressed form of Rac1 is constitutively active is in quotes to show that it is a comment made by the authors but not demonstrated in the paper. The mutation "Q61L" is information that allows us to compare the use of the construct with other experiments that use the same construct.
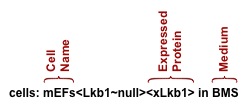
The second line supplies the name of the cells used in the experiment, any attributes, and the medium used during the experiment. In the example above the cells are mEFs (mouse embryo fibroblasts) from Lkb1 knockout mice (Lkb1~null) which have been reconstituted with exogenous Lkb1 (xLkb1). The medium used in the experiment is "BMS" which stands for Basal Medium containing Serum.
Definitions of abbreviations used in datums can be found in the Glossary of the Datum Dictionary.
The Vocabulary
The key to making a datum readable by both humans and machines is the use of a controlled vocabulary. Each of the elements described above contains categories which in turn contain lists of acceptable terms. The present set of datums was used to provide evidence for the PL STM model so the vocabulary uses that same terms as the reactions in the model. Each component has only one PL name. Names are chosen to be familiar to cellular biologists but stripped of computer confounding punctuation and alphabets such as greek. We have provided a Datum Dictionary to aid in the interpretation of a datum. A rationale for the choice of names is below.
Proteins: A protein is defined as a gene product and is commonly given a name based on the HGNC gene symbol. Peptides, splice variants, families, composites are also included under the extended Protein classification. The Protein section of the Datum Dictionary links families to their members and composites to their subunits.
Genes: In Pathway Logic, gene names are the protein name followed by "-gene".
Chemicals: The only way to uniquely identify a small molecule is to draw a structure. Datums do not use structures because they are usually not provided by the source document. We have used whatever hints the curated papers supply to assign names to chemicals. Any information collected about the named chemical is provided in the Chemical section of the Datum Dictionary.
Assays: Assays have at leaast two parts - the assay name and a detection method. The assay name is an abbreviated version of the attribute measured such as "phos" for phosphorylation or "IVKA" for in vitro kinase activity. The detection method is contained in square brackets following the assay name. The definition of these terms can be found in the Glossary of the Datum Dictionary.
Cells: Cell Line names have been simplified to make them more computer compatible. Punctation and white spaces have been removed and only upper case letters and numbers are used. Primary Cells start with a lower case letter representing the source species (h, human: m, mouse; r, rat; b, bovine; c, chicken, s, sheep, rb, rabbit; x, xenopus).
Everything else: definitions of anything that is not included above can be found in the Glossary of the Datum Dictionary.
The Datum Search Interface
The web based interface can be found at http://datum.csl.sri.com.
The search interface can be used by either typing a query directly into the search box or by using the advanced search interface which can be accessed by clicking on the double arrows under the search box.
Using the Search Box
A query is one or more terms, separated by spaces. A datum will match only if it matches every query term. A term may have the following properties:
- A term prefixed with a minus sign is negated, and will only match datums where that term is not found.
- A term in quotes will match any string component of a datum, but only in its entirety. For example, "Erk" will not match "Erks" or "Erk1", but Erk (without quotes) will match both.
- A term may take the form tag:term (or -tag:term, tag:"term", -tag:"term", etc.). If so, it only searches specific fields as constrained by the tag.
- A term with no tag and no quotes will search the full text of the datum.
- Terms may include * to match any number of any characters including none.
After a tag is typed into the search box, the next letter or letters entered will cause a drop-down list to appear which contains the first 20 elements in the field starting with that letter or letters. You can either select an item from the list or continue typing.
The list of Tags includes:
- assay
- assaytype
- cells
- change
- chemical
- extratype
- gene
- cellperturbation
- protein
- subject
- treatment
- treatmenttype
- viral (looks for protein fields with the extra stipulation that the protein must be a viral protein)
Using the Advanced Interface
If you click on the double arrows under the search box an advanced seach interface will appear. The first drop-down menu contains a list of the available tags.
The second menu allows you to refine your search using the terms:
- exists
- does NOT exist
- contains
- does NOT contain
- is exactly
- is NOT exactly
Some tags have a limited choice of terms. For those tags, a third menu containing a list of choices will appear.
Additional terms can be added using the "+" button.
When you are satisfied with your query, click on Search.
Displaying the Results
The found datums can be displayed in three different ways.
The Card format displays the first line, the environment line, a line containing the treatment times if applicable, the source line, and any extras comments. Any item colored blue can be clicked on for more information. Clicking a protein will bring up a box containing the Uniprot ID with a link to the Uniprot protein record , the HGNC symbol with a link to the HGNC record, and synonyms. Clicking on a chemical will bring up a box containing synonyms, a putative activity, and a link to the PubChem record. Clicking on the PubMed ID in the source field opens the PubMed record.
The Text format displays the entire datum in the original shorthand language.
The JSON format shows the datum parsed into JSON.
The last two formats can be downloaded for computational use.
Sample Queries
Q1: What substrates have been used for an in vitro kinase assay (IVKA) assay to show if Jnk1 is activated?
In this query, the subject is Jnk1 and the assay is In Vitro Kinase Activity.
- What to do:
- Type "subject:Jnk assay:IVKA" in the search box or use the advanced interface to do it for you.
- What you get:
- The first lines of all the datums in which Jnk1 was used in an IVKA assay. The substrate can be found in brackets after "IVKA". The substrates that have been used are: Jun, Atf2, Elk1, and MBP (myelin basic protein).
Comment: Use of "Jnk1" as the subject limits the datums to those in which Jnk1 is the only member of the Jnk family measured. If you do not care which family member you want to see then use "Jnk" instead - that will also return Jnk2, Jnk3, and Jnks (the symbol for any member of the Jnk family).
Q2: What proteins have been shown to be directly phosphorylated by Mekk1?
In PL, "directly phosphorylated" is defined by a kinase assay that contains only the kinase, the substrate, and required cofactors in a cell-free reaction.
- What to do:
- Type "treatment:Mekk1 assay:phos treattype:by" in the search box or use the advanced interface to do it for you.
- What you get:
- All the datums in which something (the subject) is phosphorylated by Mekk1 in a cell free assay. Currently the list includes Erk2, Ikk1, Ikk2, Mek1, Mkk4, Stat3.
- What to do:
- Type "subject:Ikk1 assay:IVKA perturbation:Ikk1 change:detectable" in the search box.
- What you get:
- The subject of the retrieved datums will be xIkk1 (expressed Ikk1) and the assay will be IVKA (in vitro kinase activity). The results show that exogenous Ikk1 has detectable kinase activity when expressed in HEK293, HEK293T, and HELA cells.
- What to do:
- Type "assaytype:BindingAssay subject:Nss change:detectable" in the search box or use the advanced interface to do it for you.
- What you get:
- This query found 9 interaction partners for Nss from UUKV and HRTV, 2 for Nss from TOSV, 15 for Nss from SFTSV, and 13 fro Nss from RVFV.
- What to do:
- Type "cells:MCF7 treattype:itpo treatment:xMekk1" in the search box or use the advanced interface to do it for you.
- What you get:
- The results include a responses from reporter and phosphorylation assays caused by expression of wild-type or mutant (mnr) Mekk1.
- What to do:
- Type "protein:Jnk assay:IVKA extratype:reqs treatment:IL1 treattype:irt" in the search box or use the advanced interface to do it for you.
- What you get:
- All datums using Jnks, Jnk1, Jnk2, or Jnk3 as the subject of an in vitro kinase assay from experiments in which the response of cells treated with IL1 are compared with the response from cells with various proteins removed. Removal of proteins is performed using knockouts, null mutations, omission of overexpressed proteins, or over-expression of dominant-negative proteins.
- What to do:
- Type "subject:Jnk assay:phos protein:IL1" in the search box.
- What you get:
- All datums in which Jnk is phosphorylated in response to IL1. Some of the datums include results for more than one time. Time points are listed on the third line. The relative response is indicated by the number of + symbols after each time.
- What to do:
- Type "subject:Jun-gene change:increase" in the search box or use the advanced interface to do it for you.
- What you get:
- All the datums in which Jun-gene mRNA expression is increased. These datums will provide the stimuli, treatment time, and cell line used.
- What to do:
- Type "cells:HELA change:detectable assay:prot-exp subject:Myc" in the search box or use the advanced interface to do it for you.
- What you get:
- All the datums that look at Myc protein expression in HELA cells.
- What to do:
- Type "subject:Atf2 assaytype:LocationAssay change:detectable" in the search box or use the advanced interface to do it for you.
- What you get:
- All the datums in which the location of Atf2 was determined by immunohistochemistry (IHC) or fractionation followed by Western Blot.
- What to do:
- Type "subject:Traf2 assay:ubiq change:increase treattype:irt" in the search box or use the advanced interface to do it for you.
- What you get:
- Datums that show that Traf2 is ubiquitinated in response to Tnf or anti-Cd40
- What to do:
- Type "subject:Trka assay:copptby change:detectable" in the search box or use the advanced interface to do it for you.
- What you get:
- Datums in which Trka was seen to coprecipitate with another Protein
Q3: Is expressed Ikk1 constitutively active?
Some kinases are active when over-expressed in mammalian cells
Q4: What human proteins are bound by nonstructural (Nss) viral proteins?
Q5: What happens when I over-express Mekk1 in MCF7?
Q6: What proteins have been shown to be required for the activation of Jnk (Jnk1, Jnk2, Jnk3, or Jnks) kinase activity in response to IL1 (IL1a,IL1b, or IL1)?
Q7: How long should I treat cells with IL1 to maximize Jnk phosphorylation?
Q8: What stimuli turn on Jun-gene transcription?
Q9: Do untreated HELA cells express endogenous Myc?
Q11: Where is endogenous Atf2 located in untreated cells?
Q12: What ligands cause Traf2 to be ubiquitinated?
Comment: If you were interested Traf2 ubiquitination in response to over-expression of certain proteins you would use "itpo" (in the presence of) instead of "irt" (in response to).
Q13: What proteins have been shown to coprecipitate with Trka?
Comment: There are other assays that look at the interaction between two proteins. Assay:boundby yields datums in which two recombinant or purified proteins are added together in a cell-free environment. Assay:snaggdby produces datums in which recombinant or purified proteins are added to a cell lysate then removed to see what they "snagged".