|
 |
  | ARE: Activin Responsive Element
|
 |
 |
 |
  | activin-responsive Xenopus Mix.2 promoter 12374795(C)
|
 |
 |
 |
  | The A3–Luc reporter construct was generated by subcloning the region comprising the three AREs and the core promoter from the previously described A3–CAT construct (Huang et al. 1995) into the pGL2–Basic luciferase vector (Promega).
|
 |
 |
 |
  | Huang, H.-C., L.C. Murtaugh, P.D. Vize, and M. Whitman. 1995. Identification of a potential regulator of early transcriptional responses to mesoderm inducers in the frog embryo. EMBO J. 14: 5965–5973.
|
 |
 |
 |
  | A3-Lux used in 12904571 without explanation - probably A3-Luc
|
 |
 |
 |
  | used in 12193595 "provided by Dr. M. Whitman [17]" 12193595
|
 |
 |
 |
  | 17. Hayashi, H., Abdollah, S., Qiu, Y., Cai, J., Xu, Y. Y., Grinnell, B. W., Richardson, M. A., Topper, J. N., Gimbrone, M. A., Jr., Wrana, J. L., and Falb, D. (1997) Cell 89, 1165–1173
|
 |
 |
 |
  | we constructed pAR3-lux, which contains a luciferase reporter gene under the control of three AREs
linked to a basic TATA box–containing promoter. 9215638(C)
|
 |
 |
 |
  | BRE: Bmp Response Element
|
 |
 |
 |
  | a Smad1 responsive reporter [27]. 17272273(C)
|
 |
 |
 |
  | 27. Hata, A., Seoane, J., Lagna, G., Montalvo, E., Hemmati-Brivanlou, A., and Massague, J. (2000) Cell 100, 229–240 [PMID: 10660046]
|
 |
 |
 |
  | CAGA: a Smad responsive element
|
 |
 |
 |
  | a Smad3-responsive reporter [26] 17272273(C)
|
 |
 |
 |
  | 26. Dennler, S., Itoh, S., Vivien, D., ten Dijke, P., Huet, S., and Gauthier, J. M.
(1998) EMBO J. 17, 3091–3100 [PMID: 9606191]
|
 |
 |
 |
  | CAGA reporter vectors were generated using pGL3 basic plasmid
(Promega). The CAGA boxes containing oligonucleotides were cloned into the XhoI site. The sequences of the oligonucleotides cloned are:
|
 |
 |
 |
  | CAGA boxes containing oligonucleotides:
5-TCGAGAGCCAGACAAAAAGCCAGACATTTAGCCAGACAC-3
5-TCGAGAGACAGACAAAAAGACAGACATTTAGACAGACAC-3
|
 |
 |
 |
  | CAGA mutant oligonucleotide:
5-TCGAGAGCTACATAAAAAGCTACATATTTAGCTACATAC-3
3-CTCGATGTATTTTTCGATGTATAAATCGATGTATGAGCT-5
|
 |
 |
 |
  | CAGA12 (11) reporter is driven by 12 concatamerized CAGA elements previously shown to bind complexes of Smad3 and Smad4. 10601313(C)
|
 |
 |
 |
  | 12. Yingling, J. M., Datto, M. B., Wong, C., Frederick, J. P., Liberati, N. T., and Wang, X.-F. (1997) Mol. Cell. Biol. 17, 7019–7028
|
 |
 |
 |
  | DE: a Smad responsive element
|
 |
 |
 |
  | Direct Smad2/Smad4-dependent transcription was assayed using a DE-Luciferase reporter
which is activated by a complex comprising the transcription factor Mixer with activated Smad2 and Smad4 (Germain et al., 2000). 12191474(C)
|
 |
 |
 |
  | Germain, S., Howell, M., Esslemont, G.M., and Hill, C.S. (2000). Homeodomain and winged-helix transcription factors recruit activated Smads to distinct promoter elements via a common Smad interaction motif. Genes Dev. 14, 435–451.
|
 |
 |
 |
  | the E2F–luciferase reporter construct 6×E2F–Luc (Lukas et al. 1997) 10197981(M)
|
 |
 |
 |
  | Lukas, J., T. Herzinger, K. Hansen, M.C. Moroni, D. Resnitzky, K. Helin, S.I. Reed, and J. Bartek. 1997. Cyclin E-induced S phase without activation of the pRb / E2F pathway. Genes & Dev. 11: 1479–1492. 9192874
|
 |
 |
 |
  | The pGL3-TATA-6xE2F (referred to as 6xE2F-luc) and control pGL3-TATA-basic lucif-
erase reporter plasmids were kindly donated by Ali Fattaey 9192874(M)
|
 |
 |
 |
  | an artificial promoter containing six consensus E2F sites in front of the TATA box (6xE2F-luc kindly provided by Ali Fattaey, ONYX Pharmaceuticals, Richmond, CA)
|
 |
 |
 |
  | plasmids expressing either luciferase ( A2ProLuc ) under the control of a minimal
prolactin promoter with four EGR-binding sites ( Russo et al., 1993) 11111043(M)
|
 |
 |
 |
  | Russo, M.W., Sevetson, B.R., Milbrandt, J., 1995. Identification of NAB1, a repressor of NGFI-A- and Krox20-mediated transcription. Proc. Natl. Acad. Sci. U. S. A. 92, 6873–6877.
|
 |
 |
 |
  | GAS: Gamma-interferon Activated Sequence
|
 |
 |
 |
  | pGAS-Luc GAS (4×) (AGTTTCATATTACTCTAAATC)4
|
 |
 |
 |
  | the interferon gamma-activated sequence
|
 |
 |
 |
  | 5-CTAGCAGTGTTTCCCCGAAACACGCTAG-3
|
 |
 |
 |
  | 7657660(C) contains a core sequence corresponding to the GAS element found in the IRF1 promoter.
|
 |
 |
 |
  | ISRE: Interferon Stimulated Response Element
|
 |
 |
 |
  | ISRE-Luc (Torchia et al. 1997; Nusinzon and Horvath 2003) 19171783(C)
|
 |
 |
 |
  | Torchia, J., Rose, D.W., Inostroza, J., Kamei, Y., Westin, S., Glass, C.K., and Rosenfeld, M.G. 1997. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature 387(6634): 677-684.
|
 |
 |
 |
  | Nusinzon, I. and Horvath, C.M. 2003. Interferon-stimulated transcription and innate antiviral immunity require deacetylase activity and histone deacetylase 1. Proc Natl Acad Sci U S A 100(25): 14742-14747.
|
 |
 |
 |
  | The 4X IRF3 construct contains four copies of the IRF3-binding positive regulatory domain (PRD)I/III motif of the IFNb promoter in front of a luciferase reporter gene. 15178328(C)
|
 |
 |
 |
  | IRF3-dependent luciferase reporter construct, pGL3–561 (29), was a kind gift from Dr. Ganes Sen. 18458086(C)
|
 |
 |
 |
  | 29. Grandvaux, N., Servant, M. J., tenOever, B., Sen, G. C., Balachandran, S., Barber, G. N., Lin, R., and Hiscott, J. (2002) J. Virol. 76, 5532–5539
|
 |
 |
 |
  | ISRE1: +1896 CAGTTTCCTTCTCT +1910
|
 |
 |
 |
  | ISRE2: +6670 GGAGAACAGAAACTG +6684
|
 |
 |
 |
  | ISRE1: +825 CGGTTTCTTTTGCC +839
|
 |
 |
 |
  | ISRE2: -1394 TGGAGAGAGAAACTG -1380
|
 |
 |
 |
  | a Wnt-mediated lymphoid enhancer factor-1 17272273(C)
|
 |
 |
 |
  | The -73Col-Luc reporter plasmid contains the luciferase gene under control of a truncated collagenase I promoter with a single AP-1 consensus sequence [13] 9732876(C)
|
 |
 |
 |
  | 13. Angel, P. et al. Phorbol ester-inducible genes contain a common cis element recognized by a TPA modulated trans-acting factor. Cell 49, 729–739(1987).
|
 |
 |
 |
  | contains the minimal mouse MT-I promoter (-42 to +62) 11923282(M)
|
 |
 |
 |
  | All MT-I-CAT fusions were cloned into pCATm between the SacII and HindII sites.
The MCR of pCATm contained from the T7 promoter to the Sail site of Bluescript KS+ and maintained the Sail and XbaI sites of authentic pCAT basic.
MT-I promoter constructs were generated by PCR using oligos with 5' MT-I promoter boundaries corresponding to the positions indicated by the construct name. 7800494(M)
|
 |
 |
 |
  | contains the region of the mMT-1 gene from -153 to +62
|
 |
 |
 |
  | All MT-I-CAT fusions were cloned into pCATm between the SacII and HindII sites.
The MCR of pCATm contained from the T7 promoter to the Sail site of Bluescript KS+ and maintained the Sail and XbaI sites of authentic pCAT basic.
MT-I promoter constructs were generated by PCR using oligos with 5' MT-I promoter boundaries corresponding to the positions indicated by the construct name. 7800494(M)
|
 |
 |
 |
  | contains the mouse MT-1 promoter spanning bases -264 to +43 relative the the transcription start site 18605988(M)
|
 |
 |
 |
  | cloned into pGL4.12[Luc2CP] from Promega
|
 |
 |
 |
  | mMTI-Luc was constructed by inserting a blunt-ended fragment encompassing the promoter (-727 to -13) of the mMTI gene (23) into the SmaI site of the pGL3 basic vector (Promega). 11306562(M)
|
 |
 |
 |
  | 23. Glanville, N., Durnam, D. M., and Palmiter, R. D. (1981) Nature 292, 267–269
|
 |
 |
 |
  | Mt2a-gene promo reporters
|
 |
 |
 |
  | The -358 to +40 region of the human MT2A promoter was subcloned into the vector pBlueTOPO (Invitrogen Corp.), upstream of the b-galactosidase reporter gene, as desribed previously(22). 18647441(C)
|
 |
 |
 |
  | 22. Helston RM, Phillips SR, McKay JA, et al. (2007) Zinc transporters in the mouse placenta show a coordinated regulatory response to changes in dietary zinc intake. Placenta 28, 437–444. [16914197]
|
 |
 |
 |
  | The region -358 to +40, relative to the start of transcription at +1, of the human MT2a gene was amplified by PCR from human genomic DNA (forward primer sequence: GCTCAGGTTCGAGTACAGGACAG; reverse primer sequence: CTAGAGTCGGGACAGGTTGCAC) and subcloned into the vector pBlue-TOPO (Invitrogen) upstream of the b-galactosidase reporter gene to produce the plasmid pBlue-MT2a.
|
 |
 |
 |
  | pNF-κB-Luc NF-κB (5×) (TGGGGACTTTCCGC)5 Stratagene
|
 |
 |
 |
  | used without ref in 9660950(C)
|
 |
 |
 |
  | "contains five tandem copies of the Nfkb-site from the IP10 (Cxcl10) gene" 11733537(C)
|
 |
 |
 |
  | gift from Bryan Williams (Cleveland Clnic Foundation)
|
 |
 |
 |
  | CTCAACAGAG(GGGACTTTCC)GAGAGGCCAT
|
 |
 |
 |
  | "contains Nfkb site found in the HIV long terminal repeat" 14636585(C)
|
 |
 |
 |
  | bound by Rela (supershift) in the presence of Tnf (15 min), no untreated control, 14636585(D)
|
 |
 |
 |
  | cells: mEFs on gelatin in BMS, nuclear fraction
|
 |
 |
 |
  | bound by Nfkb1 (supershift) in the presence of Tnf (15 min), no untreated control, 14636585(D)
|
 |
 |
 |
  | cells: mEFs on gelatin in BMS, nuclear fraction
|
 |
 |
 |
  | bound by Rela (supershift) in the presence of Tnf (120 min), no untreated control 14636585(D)
|
 |
 |
 |
  | cells: Mekk3-null-mEFs on gelatin in BMS, nuclear fraction
|
 |
 |
 |
  | cells: Ikba-null-mEFs on gelatin in BMS, nuclear fraction
|
 |
 |
 |
  | bound by Nfkb1 (supershift) in the presence of Tnf (120 min), no untreated control 14636585(D)
|
 |
 |
 |
  | cells: Mekk3-null-mEFs on gelatin in BMS, nuclear fraction
|
 |
 |
 |
  | cells: Ikba-null-mEFs on gelatin in BMS, nuclear fraction
|
 |
 |
 |
  | GAGCAGA(GGGAAATTCC)GTAACTT
|
 |
 |
 |
  | "contains Nfkb binding site from the IP10 (Cxcl10) gene" 11733537(C)
|
 |
 |
 |
  | CGTGGATATTCCCGGGAAAGTTTTTGGATG
|
 |
 |
 |
  | Elam1-gene promoter positions (-129 to -100)
|
 |
 |
 |
  | bound by (supershift) Rela and Nfkb1 in the presence of Tnf (3 hr), no untreated control, 7520526-Fig-3b
|
 |
 |
 |
  | not bound by (supershift) Nfkb2, Rel, Ets1, Ets2, Gabpa, BabpbS, or Arf2 in the presence of Tnf (3 hr), no untreated control, 7520526-Fig-3b
|
 |
 |
 |
  | *** rRela is BoundBy[EMSA] DNA-oligo
*** cells: none
*** oligo: CGTGGATATTCCCGGGAAAGTTTTTGGATG
*** source: 7520526-Fig-4b
|
 |
 |
 |
  | *** rNfkb1 is BoundBy[EMSA] DNA-oligo
*** cells: none
*** oligo: CGTGGATATTCCCGGGAAAGTTTTTGGATG
*** source: 7520526-Fig-4b
|
 |
 |
 |
  | reporter constructs containing repeated IRF binding sites; regulated by multimerized IRF binding sites 16116171(C)
|
 |
 |
 |
  | Pai1 promoter was provided by David Loskutoff 10085121(C)
|
 |
 |
 |
  | -2329 to +510 of the Myc-gene promoter in pGL3-basic 15684397(C)
|
 |
 |
 |
  | contains the Tgfb1 responsive element of PAI1 promoter (nucleotides -740 to -636) and the promoter region of the hujan interferon-beta gene from nucleotides -55 to +19 [16]. 8533096
|
 |
 |
 |
  | 16. R Hirai and T Fujita, personal communication
|
 |
 |
 |
  | 14525954(M): gift from X-Ceptor Therapeutics
|
 |
 |
 |
  | PPRE-tk-LUC reporter that contains three copies of a consensus binding site cloned upstream of the heterologous herpes simplex virus tk promoter linked to the LUC reporter gene.
|
 |
 |
 |
  | Pten-gene promo reporters
|
 |
 |
 |
  | The human PTEN gene promoter-luciferase reporter construct was a gift from Eileen Adamson (Burnham Institute, La Jolla, Calif.). The PTEN-Luc reporter construct was made by subcloning the 1,978-bp genomic DNA region upstream of the human PTEN gene into the empty pGL3 basic-Luc vector (47).
|
 |
 |
 |
  | 47. Virolle, T., E. D. Adamson, V. Baron, D. Birle, D. Mercola, T. Mustelin, and I. de Belle. 2001. The Egr-1 transcription factor directly activates PTEN during irradiation-induced signalling. Nat. Cell Biol. 3:1124–1128. 11781575
|
 |
 |
 |
  | We amplified 5′ PTENregulatory sequences by PCR from genomic DNA using a pair of appropriate primers (5′-KpnI-GCCGGGTTTCACGCGGC-3′ and 5′-HindIII-GTCTGGGAGCCTGTGG-3′) located respectively at the position −1 and −1,978 from the ATG.The amplified product was purified and cloned into the KpnI/HindIII-digested pGL3 basic reporter gene to give the PTEN–luc construct. 11781575(M)
|
 |
 |
 |
  | Because the PTEN promoter contains two potential NFKB binding sites (GGGAATCTCT at nucleotide position -1565 and GGGTATTCCC at nucleotide position -1441) in
the far upstream region, we made a 600-bp deletion in the PTEN promoter to eliminate both putative NFKB binding sites (14729949-Fig. 4A).
|
 |
 |
 |
  | pTOPFLASH Millipore Catalog # 21-170
|
 |
 |
 |
  | three copies of the optimal Tcf motif CCTTTGATC upstream of a minimal c-Fos promoter driving luciferase expression 9065401(M)
|
 |
 |
 |
  | a synthetic TCF4 (Tcf7l2) protomoter
|
 |
 |
 |
  | TCF binding site (wt) sequence:
AAGATCAAAGGGGGTAAGAKCAAAGGGGGTAAAATCAAAGGGGGCCCCCTTTGATCTTACCCCCTTTGATCTTACCCCCTTTGATCTTA
|
 |
 |
 |
  | used in 15684397 15026412 9065401 12727872
|
 |
 |
 |
  | SBE: Smad Binding Element
|
 |
 |
 |
  | The new reporters described here were derived from the pGL3 promoter luciferase vector (Promega). Inserts containing SBE sequences were generated by ligation of concatamerized oligonucleotides to KpnI-digested pGL3 (Hermeking et al., 1997), generating the vector pSBE4-luc (four copies of the SBE). The complementary oligonucleotides used for pSBE4-luc construction were 5-TAAGTCTAGACGGCAGTCTAGACGTAC-3 and 5-GTCTAGACTGCCGTCTAGACTTAGTAC-3,
|
 |
 |
 |
  | Plasmid SBE-luc contains the luciferase gene under control of the Smad-binding elements (SBE) (35) 12621041
|
 |
 |
 |
  | SBE-luc contains four copies of SBE linked to a luciferase reporter gene (35).
|
 |
 |
 |
  | this paper only has 33 references
|
 |
 |
 |
  | We used the (SBE)4-luciferase reporter, in which transcription is directed from four tandem Smad-binding elements, to evaluate the activity of Smad4 as a coactivator (35). 12740389(C)
|
 |
 |
 |
  | 35. Zawel, L., Dai, J. L., Buckhaults, P., Zhou, S., Kinzler, K. W., Vogelstein, B. and Kern, S. E. (1998) Mol. Cell 1, 611–617 9660945
|
 |
 |
 |
  | the 4xSBE (Smad-binding element) promoter (37).
|
 |
 |
 |
  | 37. Zawel, L., Dai, J. L., Buckhaults, P., Zhou, S., Kinzler, K. W., Vogelstein, B. and Kern, S. E. (1998) Mol. Cell 1, 611–617 9660945
|
 |
 |
 |
  | co-transfected with the SBE4-Luc reporter gene, which contains four copies of the Smad-binding element. (no refs provided)
|
 |
 |
 |
  | SBE4-luc, which contains four tandem repeats of the Smad-binding element (SBE) (27).
|
 |
 |
 |
  | 27. Zawel, L., Dai, J. L., Buckhaults, P., Zhou, S., Kinzler, K. W., Vogelstein, B.,
and Kern, S. E. (1998) Mol. Cell 1, 611–617 9660945
|
 |
 |
 |
  | we used (SBE)4-Luc, because TGF-b-induced activation of (SBE)4-Luc, driven by four repeats of the CAGACA sequence identified as Smad binding element in the JunB promoter (Jonk et al., 1998), was dependent on expression of Smad3, but not on expression of Smad2 (Piek et al., 2001).
|
 |
 |
 |
  | Jonk LJ, Itou S, Heldin CH, ten Dijke P and Kruijer W. (1998). J. Biol. Chem., 273, 21145–21152.
|
 |
 |
 |
  | artificial Smad2/3-responsive promoter (SBE4X-luciferase)
|
 |
 |
 |
  | a mammalian TGfbeta-inducible luciferase reporter construct (SBE-4X) as described previously (19).
|
 |
 |
 |
  | 19. Seoane, J., Le, H. V., Shen, L., Anderson, S. A., and Massague´, J. (2004) Cell
117, 211–223
|
 |
 |
 |
  | Smad-binding element (SBE)2-Luc (39).
|
 |
 |
 |
  | the Smad3-specifc SBE2-Luc luciferase reporter (39)
|
 |
 |
 |
  | 39. Zawel, L., Dai, J. L., Buckhaults, P., Zhou, S., Kinzler, K. W., Vogelstein, B., and Kern, S. E. (1998) Mol. Cell 1, 611–617.
|
 |
 |
 |
  | SIE: Stat inhibitory element
|
 |
 |
 |
  | The 1X SIE-CAT (TTCCCGTCAA) contains the sequence of the human c-fos promoter spanning from nucleotides -350/-336 inserted into a pBLCAT5 derived plasmid.
|
 |
 |
 |
  | SRE: Serum Response Element
|
 |
 |
 |
  | CC(A/T)6GG CCWWWWWWGG 11029289(C)
|
 |
 |
 |
  | SRF-dependent element 11598209(C)
|
 |
 |
 |
  | SrfRE: Srf Response Element
|
 |
 |
 |
  | Stat3RE: Stat3 Response Element
|
 |
 |
 |
  | The Stat3 luciferase reporter gene plasmid was kindly provided by Dr. Geeta Devgan (The Rockefeller University).
|
 |
 |
 |
  | a luciferase reporter gene that contained four copies of the Stat3 binding site fused to a minimal promoter
|
 |
 |
 |
  | Stratgene cis-reporter plasmids
|
 |
 |
 |
  | Plasmid configuration sequence
|
 |
 |
 |
  | pC/EBP-Luc C/EBP (3×) (ATTGCGCAAT)3
|
 |
 |
 |
  | the CCAAT-enhancer binding protein (C/EBP) transcription recognition sequences
|
 |
 |
 |
  | pCRE-Luc CRE (4×) (AGCCTGACGTCAGAG)4
|
 |
 |
 |
  | the consensus sites for cyclic AMP response element
|
 |
 |
 |
  | pCRE-hrGFP CRE (4×) (AGCCTGACGTCAGAG)4
|
 |
 |
 |
  | the consensus sites for cyclic AMP response element
|
 |
 |
 |
  | pDR1-Luc DR1 (5×) (AGGTCAN)5
|
 |
 |
 |
  | the retinoid X receptor response element (DR1/RXRE)
|
 |
 |
 |
  | pDR3-Luc DR3 (4×) (AGGTCANNN)4
|
 |
 |
 |
  | the vitamin D receptor response element (DR3/VDR)
|
 |
 |
 |
  | pDR4-Luc DR4 (2×) (CAGGAGGTCA)3
|
 |
 |
 |
  | the active thyroid hormone receptor response element (DR4/TR)
|
 |
 |
 |
  | pDR5-Luc DR5 (5×) (AGGTCANNNNN)5
|
 |
 |
 |
  | the retinoic acid receptor response element (DR5/RARE)
|
 |
 |
 |
  | pGRE-Luc GRE (4×) (GGTACATTTTGTTCT)4
(second repeat is in antisense orientation)
|
 |
 |
 |
  | the glucocorticoid response element (GRE)7
|
 |
 |
 |
  | pISRE-Luc ISRE (5×) (TAGTTTCACTTTCCC)5
|
 |
 |
 |
  | the interferon-stimulated response element
|
 |
 |
 |
  | pLILRE-Luc LILRE (4×) (TCACTTCCTGAGAG)4
|
 |
 |
 |
  | the LPS IL-1β response element
|
 |
 |
 |
  | pNFAT-Luc NFAT binding site (4×) (GGAGGAAAAACTGTTTCATACAGAAGGCGT)4
|
 |
 |
 |
  | the nuclear factor of activated T cells transcription recognition sequences
|
 |
 |
 |
  | pNFAT-hrGFP NFAT binding site (4×) (GGAGGAAAAACTGTTTCATACAGAAGGCGT)4
|
 |
 |
 |
  | the nuclear factor of activated T cells transcription recognition sequences
|
 |
 |
 |
  | pNF-κB-hrGFP NF-κB (5×) (TGGGGACTTTCCGC)5
|
 |
 |
 |
  | the binding sites for nuclear factor κB
|
 |
 |
 |
  | pTARE-Luc TARE (3×) (TCTCAATCCACAATCTCGGAGTATGTCTAGACTGACAATG)3
|
 |
 |
 |
  | the TGF-β/Activin response element
|
 |
 |
 |
  | TIE: Tgfb Inhibitory Element
|
 |
 |
 |
  | consensus sequence: 5'-GNNTTGGtGa-3'
|
 |
 |
 |
  | A TGF-beta inhibitory element (TIE) was first characterized as essential to mediating TGF-beta repressive effects in the promoter of the TGF-beta-repressed gene stromelysin 1 (39). The identified stromelysin 1 TIE was compared to the promoter sequence of other TGF-beta-repressed genes, such as collagenase 1/matrix metalloproteinase 1 (MMP-1), urokinase, and c-myc, and the following nucleotides, in which uppercase letters signify invariance and lowercase letters signify preferred sequence, became known as a consensus TIE: 5'-GNNTTGGtGa-3' (39).
|
 |
 |
 |
  | 39. Kerr, L. D., D. B. Miller, and L. M. Matrisian. 1990. TGF-beta 1 inhibition
of transin/stromelysin gene expression is mediated through a Fos binding
sequence. Cell 61:267–278.
|
 |
 |
 |
  | TIEs within the rat stromelysin and rabbit MMP-1 promoters have been shown to bind TGF-beta-induced c-Fos, and this mechanism of c-Fos upregulation and subsequent TIE binding has been reported as essential in mediating the transcriptional repression of these promoters by TGF-beta (39, 82).
|
 |
 |
 |
  | 82. White, L. A., T. I. Mitchell, and C. E. Brinckerhoff. 2000. Transforming growth factor beta inhibitory element in the rabbit matrix metalloproteinase-1 (collagenase-1) gene functions as a repressor of constitutive transcription. Biochim. Biophys. Acta 1490:259–268.
|
 |
 |
 |
  | Although the recruitment of c-Fos to the putative c-myc TIE (5'-GGCTTGGCGG-3'; nucleotides -84 to -75 relative to the major P2 transcriptional start site) has not been implicated in the TGF-beta-mediated repression of c-myc, an overlapping, consensus E2F binding site (5'-GGCGGGAAA-3'; -79 to -71) has been shown to be important for transactivation of the c-myc promoter by E2F and Ets transcription factors (33, 67).
|
 |
 |
 |
  | 33. Hiebert, S. W., M. Lipp, and J. R. Nevins. 1989. E1A-dependent transactivation of the human MYC promoter is mediated by the E2F factor. Proc. Natl. Acad. Sci. USA 86:3594–3598.
|
 |
 |
 |
  | 67. Roussel, M. F., J. N. Davis, J. L. Cleveland, J. Ghysdael, and S. W. Hiebert. 1994. Dual control of myc expression through a single DNA binding site targeted by ets family proteins and E2F-1. Oncogene 9:405–415.
|
 |
 |
 |
  | TRE: TPA Response Element
|
 |
 |
 |
  | TRE-Luc contains four tandem copies of the AP-1 consensus sequence in front of the luciferase gene [12]. 9732876(C)
|
 |
 |
 |
  | 12. Renshaw, M.W., McWhirter, J.R.&Wang, J.Y. The human leukemia oncogene bcr-abl abrogates the anchorage requirement but not the growth factor requirement for proliferation. Mol. Cell. Biol. 15, 1286–1293(1995).
|
 |
 |
 |
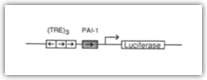
  | 1333888(C): The 3TP promoter contains three consecutive TPA response elements (TREs) and a portion of the plasminogen activator inhibitor 1 (PAI-1) promoter region (Figure 1A) . The choice of TRE concatemers and the PAI- promoter was based on the responsiveness of these elements in TGFR (de Grnnt and Kruijer, 1990; Keeton et al., 1991). Other details of the design and properties of TGF6 reporter systems will be presented elsewhere (Juan Carcamo and Joan Massague, unpublished data).
|
 |
 |
 |
  | Briefly, a modified plasmid (pG5E4T) containing adenovirus E4 promoter sequences from positions -38 to +36 (Lin et al., 1988) was used to construct the 3TP promoter. A TGF6 responsive element (Keeton et al., 1991) from positions -636 to -740 in the human PAI- promoter was synthesized and inserted upstream of the adenovirus E4 sequences. The TRE corresponding to the -73 to -42 region of the human collagenase gene was multimerized (de Groot and Kruije, 1990) and inserted upstream of the PAI- sequence generating the 3TP promoter. A fragment (Xhol-EcoRI) containing the 3TP promoter was isolated and cloned into Xhol-Hindlll-digested luciferase expression vector (pGL2, basic vector; Promega) generating p3TP-Lux.
|
 |
 |
 |
  | de Groot, R. P., and Kruijer, W. (1990). Transcriptional activation by TGF51 mediated by the dyad symmetry element (DSE) and the TPA responsive element (TRE). Biochem. Biophys. Res. Commun. 168, 1074-1081.
|
 |
 |
 |
  | Keeton, M. R., Curriden, S. A., van Zonneveld, A. J., and Loskutoff, D. (1991). Identification of regulatory sequences in the type 1 plasminogen activator inhibitor gene responsive to transforming growth factor beta. J. Biol. Chem. 266, 23046-23052.
|
 |
 |
 |
  | Lin, Y.-S., Carey, M. F., Ptashne, M., and Green, M. R. (1988). GAL4 derivatives function alone and synergistically with mammalian activators in vitro. Cell 64, 659-664.
|
 |
 |
 |
  | 9215638(C): contains elements from the PAI-1 promoter, is TGF responsive, and drives expression of a luciferase reporter gene (Wrana et al., 1992)
|
 |
 |
 |
  | Wrana, J.L., Attisano, L., Carcamo, J., Zentella, A., Doody, J., Laiho, M., Wang, X.-F., and Massague ́ , J. (1992). TGF- signals through a heteromeric protein kinase receptor complex. Cell 71, 1003–1014 1333888
|
 |
 |
 |
  | 11356828(C) "3TP-Lux reporter was kindly provided by Dr J Massague"
|
 |
 |
 |
  | contains both PAI-1 and collagenase promoter elements (31)
|
 |
 |
 |
  | 31. Wrana, J. L., Attisano, L., Carcamo, J., Zentella, A., Doody, J., Laiho, M.
Wang, X. F., and Massague´ , J. (1992) Cell 71, 1003–1014
|
 |
 |
 |
  | 12379659(C): p3TP-Lux contains three AP-1 sites and the plasminogen activator inhibitor-1 (PAI-1) promoter (30).
|
 |
 |
 |
  | 30. Lagna, G., Hata, A., Hemmati-Brivanlou, A., and Massague, J. (1996) Nature 383,
832–836
|
 |
 |
 |
  | Stratagene #219073 (AP-1 cis-Reporting System)
|
 |
 |
 |
  | The reporter plasmid 5xjun2-tata-luciferase was constructed by introduction of the XhoI-StyI fragment of pGl2 into 5xjun2-tata-CAT. (8382609). [9553051]
|
 |
 |
 |
  | Contains a high-affinity c-Jun:ATF2-binding site TTACCTCA, jun2 site of the
c-jun promoter (8382609). [9553051]
|
 |
 |
 |
  | Has been shown to specifically respond to over-expressed Jun:ATF2. (8382609). [9553051]
|
 |
 |
 |
  | The jun2-luc reporter plasmid (pGL3–5xjun2-luc) was a gift from Dr Dorien Peters. [16511568]
|
 |
 |
 |
  | a reporter driven by the TGFb responsive region of the collagenase promoter, modified to contain intact AP-1 sites, but mutant SMAD-binding sites (12). 10601313(C)
|
 |
 |
 |
  | 12. Yingling, J. M., Datto, M. B., Wong, C., Frederick, J. P., Liberati, N. T., and Wang, X.-F. (1997) Mol. Cell. Biol. 17, 7019-7028
|
 |
 |
|
 |